Method for extracting microorganism total DNA from coal seam water sample
A technology for microorganisms and water samples, applied in the field of molecular biology, can solve the problems of complex physical and chemical properties, unsuitable microbial genomes, and complex composition of coal seam water samples, achieving the effect of good integrity and high purity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] (1) Take 15mL coal seam water sample and centrifuge at 1500×g for 5min, collect the supernatant in a serum bottle, use suction filtration to collect the microorganisms in the water sample onto the filter membrane, then break the filter membrane into pieces and put them into 5mL STE Buffer (NaCl0.12g, 20mM Tris-HCl10mL, 0.5MEDTA40μL, ddH 2 (09960 μL, to a final volume of 20 mL), transfer to a pre-cooled screw-cap tube containing 0.6 g of glass beads with a diameter of 0.3 mm.
[0031] (2) Break it with a bead mill Mini-BeadBeater (MBB), run it at 2500rpm for 30s, then put the screw cap tube on ice, take the supernatant and transfer it to a new 2mL Ep tube, fill the screw cap tube with STE again, Run at 2500rpm for 30s, let stand on ice, and combine with the first supernatant.
[0032] (3) Add 24 μL of proteinase K (20 mg / mL) and 216 μL of 10% SDS in a 55° C. water bath to continue lysing and digesting the bacteria for 3 h.
[0033](4) Add 1 mL of phenol-chloroform-isoa...
Embodiment 2
[0042] (1) Collection of bacteria: Take 6 mL of coal anaerobic fermentation broth and put them into sterile Ep tubes, and collect the bacteria by filtration with a 0.22 μm membrane. After the membrane is cut into pieces, take a part of the filter membrane and place it in 2mL of STE buffer, move it to a pre-cooled screw cap tube containing about 0.6g of glass beads with a diameter of 0.3mm, and extract the total DNA with the method of the present invention; the other part contains The filter membrane of the bacteria was used as a control to extract the total DNA with a kit from a certain company.
[0043] (2) Break the cells with MBB, the operating program is 2500rpm, 30s, put the screw-cap tube on ice after breaking, take the supernatant and transfer it to a new 2mLEP tube, fill the screw-cap tube with STE buffer again, Run at 2500rpm for 30s, and combine the supernatant with the first supernatant.
[0044] (3) Add 18 μL of proteinase K (20 mg / mL) and 180 μL of 10% SDS in a 5...
Embodiment 3
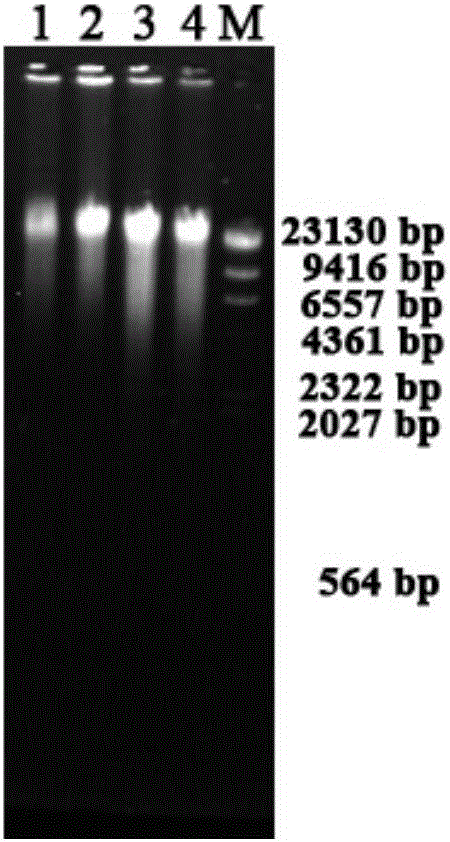
[0054] Using the genomes extracted in Examples 1 and 2 as templates, the bacterial 16S rDNA gene V1-V3 regions and the archaeal 16S rDNA gene V3-V4 region fragments were respectively amplified for subsequent high-throughput sequencing. The PCR primers were bacterial 27F (AGAGTTTGATCCTGGCTCAG) / 518R (ATTACCGCGGCTGCTGG), the PCR reaction system was 50 μL, including 1 μL of genomic DNA, 5 μL of 10×EasyTaqbuffer, 4 μL of dNTPs (2.5 mM), 2 μL of upstream and downstream primers (10 μM), 1 μL of EasyTaqPloymerase, and ddHO 2 O was added to 50 μL, and the PCR amplification program was pre-denaturation at 95°C for 5 minutes, followed by 30 cycles: 95°C for 30 s, 55°C for 30 s, 72°C for 45 s, and final extension at 72°C for 10 min. The PCR product was detected by 2% agarose gel electrophoresis, and the results were as follows: Figure 4 As shown, because the total DNA quality of the microorganisms extracted by the kit method is general, the situation that the bacterial 16SrDNA fragments ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com