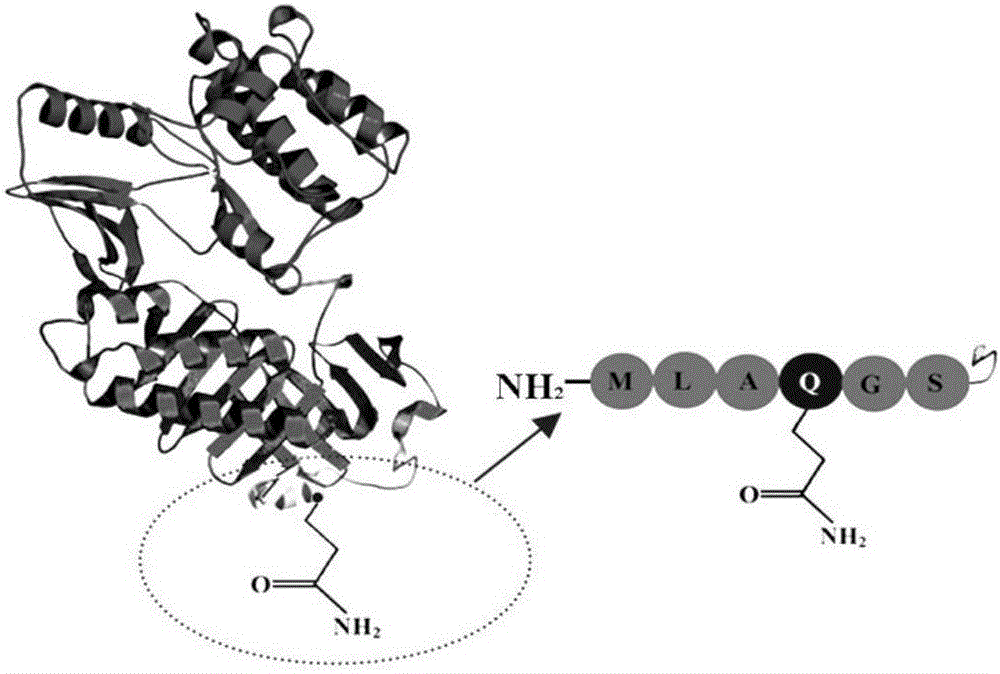
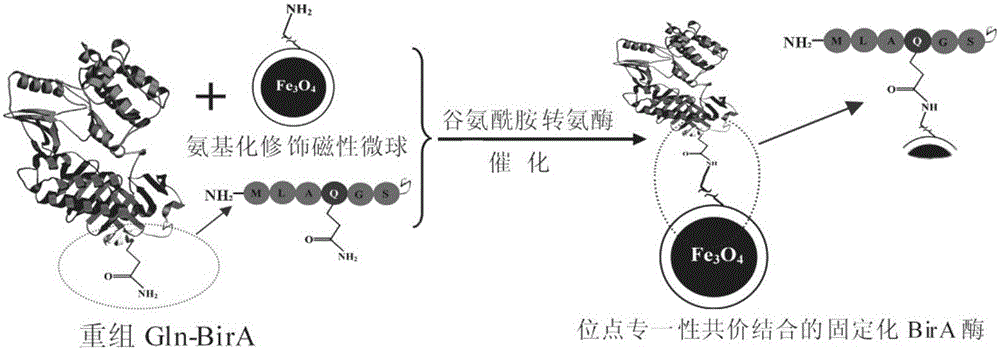
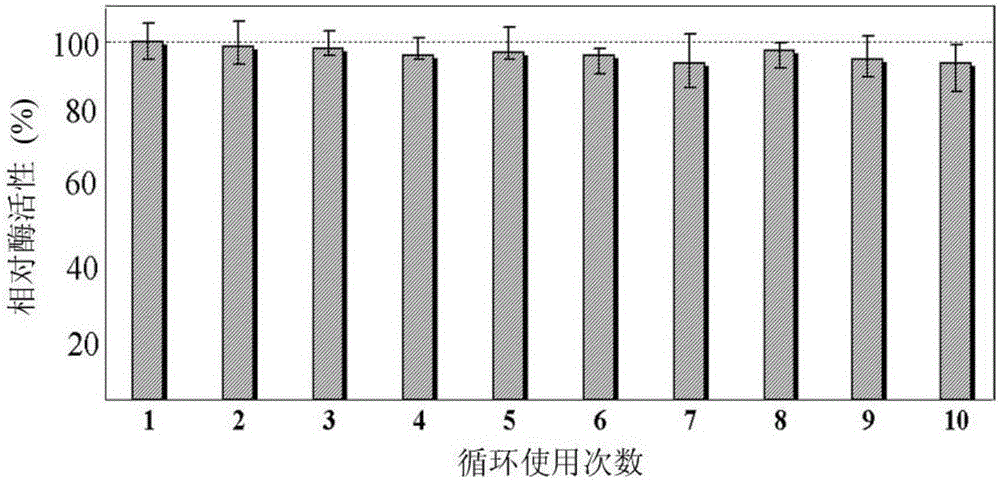
Directional covalent immobilization method of biotin-protein ligases (BirA) on magnetic microsphere surface
A biotin ligase, magnetic microsphere technology, applied in the field of biochemistry, can solve the problems of no fixed binding mode, affecting the catalytic activity of the enzyme, position uncertainty, etc., to avoid random coupling and improve the use efficiency of enzymes , the effect of high fixed amount
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Embodiment 1, the construction of recombinant MLAQGS-His-BirA gene prokaryotic expression vector
[0032] According to the BirA gene sequence of Escherichia coli K-12MG1655 (No: EG10123) registered in GenBank, the following primers were designed:
[0033] Primer 1, as shown in SEQ ID No: 1:
[0034] 5'-GAATTCGATGCTAGCGCAGGGATCACACCATCACCATCACCATATGAAGGATAACA-3';
[0035] Primer 2, as shown in SEQ ID No: 2:
[0036] 5′-CCGGGATCCTTTATTTTTCTGCACTACGCAGGG-3′
[0037] Among them, Primer 1 contains EcoR I restriction site, MLAQGS sequence, 6×His sequence, and Primer 2 contains BamH I restriction site.
[0038] Using the E.coli DH5α genome as a template, Primer 1 and Primer 2 were used as primers to amplify the BirA gene by PCR, and the obtained gene fragment was cloned into the pUC18 plasmid EcoR I / BamH I restriction site according to molecular biology methods to obtain pMLAQGS- His-BirA expression plasmid vector.
Embodiment 2
[0039] Embodiment 2, the construction of recombinant BirA-His-MLAQGS gene prokaryotic expression vector
[0040] According to the BirA gene sequence of Escherichia coli K-12MG1655 (No: EG10123) registered in GenBank, the following primers were designed:
[0041] Primer 3, as shown in SEQ ID No: 3:
[0042] 5'-GAATTCGATGAAGGATAACA-3';
[0043] Primer 4, as shown in SEQ ID No: 4:
[0044] 5′-CCGGGATCCCTGGTGCTGGTGCTGGTGTGATCCCTGCGCTAGCATTTATTTTTCTGCACTACGCAGGG-3′
[0045] Among them, Primer 3 contains EcoR I restriction site, and Primer 4 contains 6×His sequence, MLAQGS sequence and BamH I restriction site.
[0046] Using the E.coli DH5α genome as a template, Primer 3 and Primer 4 were used as primers to amplify the BirA gene by PCR, and the obtained gene fragment was cloned into the pUC18 plasmid EcoR I / BamH I restriction site according to molecular biology methods to obtain pBirA- His-MLAQGS expression plasmid vector.
Embodiment 3
[0047] Example 3, Preparation and Purification of Genetic Engineering Technology of Recombinant MLAQGS-His-BirA and BirA-His-MLAQGS
[0048] pMLAQGS-His-BirA plasmid CaCl 2 E.coli BL21(DE3) was transformed by the method to construct pMLAQGS-His-BirA / BL21 engineering bacteria.
[0049] pBirA-His-MLAQGS plasmid CaCl 2 E.coli BL21(DE3) was transformed by the method to construct pBirA-His-MLAQGS / BL21 engineering bacteria.
[0050] The above two kinds of engineered bacteria were activated overnight, transferred to fresh LB medium (ampicillin concentration 100 μg / mL) according to 5% inoculum size, cultured with shaking at 37° C. for 12-16 hours, and centrifuged to collect the bacteria. Resuspend the bacteria in PBS solution containing 20mM imidazole and 500mM NaCl (20mM, pH 8.0), break the wall by ultrasonic, centrifuge at 10000rpm to separate the bacteria fragments, filter the supernatant through a 0.22μm filter membrane, and flow through the HisTrap chromatography column at 0.5m...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com