Kit for simultaneously detecting four pathogenic bacteria and non-diagnostic detection method thereof
A pathogenic bacteria, positive technology, applied in the field of biotechnology detection, can solve the problems of poor domestic reagents, rare detection methods, invalid detection results, etc., to avoid false negative results, high degree of automation, and improved sensitivity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
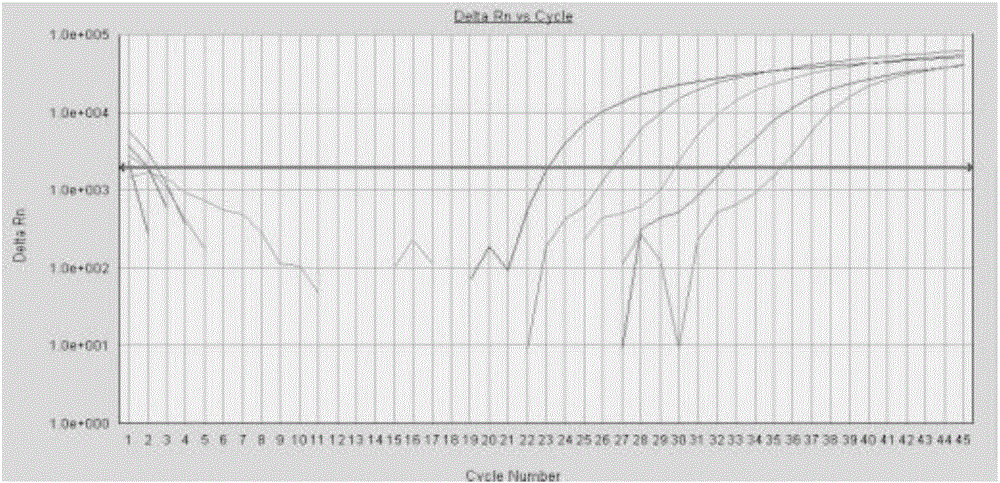
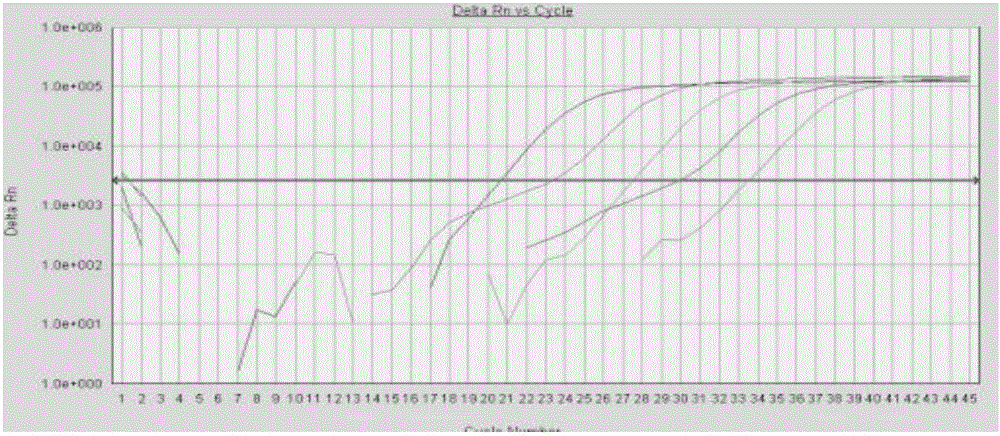
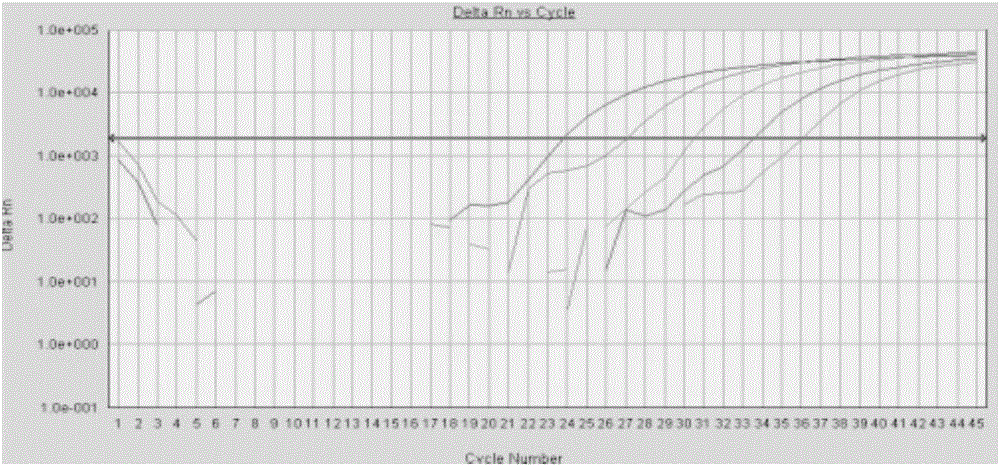
Image
Examples
Embodiment 1
[0055] The design of embodiment 1 primer, probe
[0056] Firstly, the specific target genes of the four pathogenic bacteria were screened separately. According to the purpose of detection, multiple pathogenic bacterial gene sequences were downloaded from GenBank for each pathogen, compared and analyzed, and the conserved region was selected, and Array Designer4.0 software was used to assist in the design of amplification primers and suitable Hybridization probe for fluorescent quantitative PCR reaction system.
[0057] Since the present invention is a multiplex fluorescence quantification, the selection of probe labels is more critical at first, and secondly, the probes designed by the software must be screened, and the probe signals of the four genes cannot interfere with each other, so it is necessary to consider four pairs of genes when designing the probes. The primers and the four probes cannot interfere with each other.
[0058] Fluorescent quantitative PCR detection is...
Embodiment 2
[0082] Example 2 Detection of Four Pathogenic Bacteria of Yersinia pestis, Tularemia, Burkholderia pseudomallei and Brucella by Common PCR
[0083] According to Example 1, the conserved sequence of the Pla gene fragment of Yersinia pestis was screened, and a pair of oligonucleotide sequences (primers) and probes were designed. It was finally determined that the size of the Yersinia pestis primer amplified fragment was 113bp, and the nucleotide sequence was shown in SEQ ID No.4.
[0084] SEQ ID No. 4: atgtcacacc taatgccaaa gtctttgcgg aatttacata cagtaaatatgatgagggca aaggaggtac tcagatcatt gataagaata gtggagattc tgtctctatt ggc.
[0085] The ISFtu gene was selected for the tularensis, and the size of the fragment amplified by the primers was 113bp, and the nucleotide sequence was shown in SEQ ID No.8.
[0086] SEQ ID No. 8: agctacttta ctatcatgag ttttaccttc tgacaacaat atttctattggattacctaaagcatcagtc atagcatgga ttttagtggt tatcccacca actgatctac caa.
[0087] Burkholderia pseudomallei ...
Embodiment 3
[0096] Example 3 Fluorescence quantitative PCR detection kit for four kinds of pathogenic bacteria
[0097] Fluorescent quantitative PCR detection kit for four pathogens of Yersinia pestis, Tularemia, Burkholderia pseudomallei and Brucella include the following components:
[0098] Premix EX Taq™×2;
[0099] The upstream primer sequence of Yersinia pestis is shown in SEQ ID No.1, the downstream primer sequence is shown in SEQ ID No.2, the probe sequence is shown in SEQ ID No.3, and 5 of the probe SEQ IDNo.3 'marks FAM, 3' marks BHQ1;
[0100] The upstream primer sequence of the tularemia is shown in SEQ ID No.5, the downstream primer sequence is shown in SEQ ID No.6, and the probe sequence is shown in SEQ ID No.7 wherein, the probe SEQ ID No.7 The 5' tag Texred, 3' tag BHQ2;
[0101] The upstream primer sequence of the Burkholderia pseudomallei is shown in SEQ ID No.9, the downstream primer sequence is shown in SEQ ID No.10, and the probe sequence is shown in SEQ ID No.11, ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| wavelength | aaaaa | aaaaa |
| wavelength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com