Lipase mutant deriving from talaromyces thermophilus, coding gene and application thereof
A technology for encoding genes and mutants, applied in the application field of 2-carboxyethyl-3-cyano-5-methylhexanoic acid, can solve the problem of restricting large-scale industrial applications, harsh production conditions, and the amount of organic solvents used Large and other problems, to achieve the effect of good application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Example 1 Construction of Lipase Mutation Library
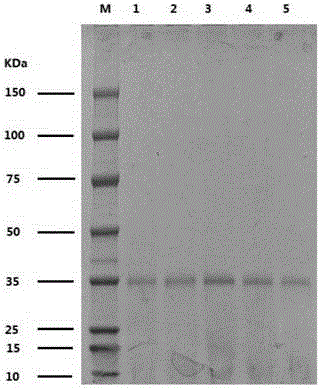
[0031] The pET-28b-TTL plasmid, which was cloned from the lipase gene of Talaromyces thermophilus (GenBank accession No.JF414585.1) and linked to pET-28b(+) as a template (nucleotide sequence SEQ ID NO.2 ), using T7promoter and T7terminator as primers (Table 1) for PCR amplification, and randomly introducing mutations. PCR reaction system (50μL): template 0.5~20ng, 1×Taq Buffer (without Mg 2+ ), 0.2mM dGTP, 0.2mM dATP, 0.1mM dCTP, 0.1mM dTTP, 0.2mMMnCl 2 , Primers T7promoter and T7terminator each 0.2μM, Taq DNA polymerase 5U. PCR conditions (1) 95°C pre-denaturation for 3 minutes; (2) 95°C denaturation for 30s; (3) 55°C annealing for 30s; (4) 72°C extension for 1 minute, steps (2) to (4) total 30 cycles; (5) ) Finally, extend at 72°C for 10 minutes and store at 4°C. After the PCR product was analyzed by agarose gel electrophoresis, the gel was cut and recovered, and it was digested with NcoI and Xhol, ligated with the s...
Embodiment 2
[0034] Example 2 High-throughput screening of mutants
[0035] Pick the single colony in Example 1 and culture it in a 96 deep-well plate, add 200μL LB medium (containing 50μg / mL kanamycin) to each well plate, and incubate at 37°C for 2.5h to OD 600 When it is about 0.6, add the final concentration of 0.1mM IPTG to induce, and induce culture at 28℃ for 20h. Centrifuge the bacteria in the 96-deep well plate for 15 min (5000 rpm, 4°C), discard the supernatant, and resuspend the bacteria in Tris-HCl (20 mM, pH 8.0) buffer as the biocatalyst for the reaction. The bacterial suspension was added to each well of a 96 shallow-well plate, 200 μL per well, then the substrate CNDE (final concentration 100 mM) was added, and 0.01% bromothymol blue was used as an indicator, and reacted at 30°C for 1-3 hours.
[0036] According to the speed of the color change, the bacteria with increased vitality were selected (using the wild-type strain as the control), and the catalytic activity of CNDE was v...
Embodiment 3
[0037] Example 3 Site-directed mutation and screening
[0038] In order to further improve the enzyme activity of the mutants, site-directed mutagenesis is used to continue to screen for better complex mutants. Using the expression plasmid pET-28b-L259F as a template, site-directed mutagenesis was performed through whole plasmid amplification. The PCR system (50μL) is: template 0.5-20ng, 5×PrimeSTAR Buffer 10μL, dNTP Mixture 4μL, mutation primer (Table 1) each 1μL, PrimeSTAR DNA Polymerase 0.5μL. PCR conditions (1) 95°C pre-denaturation for 3 minutes; (2) 95°C denaturation for 15 seconds; (3) 55°C annealing for 10 seconds; (4) 72°C extension for 5 minutes, steps (2) to (4) total 30 cycles; (5) ) Finally, extend at 72°C for 10 minutes and store at 4°C. The PCR product was verified by agarose gel electrophoresis, digested with DpnI, and then introduced into E.coli BL21(DE3), and spread on an LB plate containing 50μg / mL kanamycin. The catalytic activity of each mutant on CNDE was...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com