Acidic lipoxygenase and preparation method and application thereof
A lipoxygenase, acid technology, applied in the field of acid lipoxygenase and its preparation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Example 1: Myxococcus xanthus Cloning of DK1622 lipoxygenase gene (rMxLOX)
[0029] centrifuge collection Myxococcus xanthus DK1622 cells, extracted with Shanghai Sangon Genomic DNA Extraction Kit Myxococcus xanthus Genomic DNA of DK1622.
[0030] Two primers were designed according to the Myxococcus aurantiacus lipoxygenase gene (No.CP006832.1) registered in the Genebank database:
[0031] Upstream primer F-1: 5'-ATGAAACGCAGGAGTGTGCTCTTG-3' (SEQ ID NO.3);
[0032] Downstream primer R-1: 5'-TCAGATATTGGTGCTCGCCGGGATC-3' (SEQ ID NO.4);
[0033] obtained by extracting Myxococcus xanthus DK1622 genomic DNA is used as a template, and the above two primers are used for PCR amplification. The PCR reaction system is as follows:
[0034] 2× GC Buffer 25 µL MXLOX-F 2 µL MXLOX-R 2 µL 2.5 mM dNTPs 8 µL genomic DNA 1 µL Pfu DNA polymerase 1 µL wxya 2 o
Make up to 50 µL
[0035] PCR reaction conditions: pre-denaturati...
Embodiment 2
[0037] Embodiment 2: the construction of prokaryotic expression vector of myxococcus aureus DK1622 lipoxygenase gene (rMxLOX) (attachment figure 1 )
[0038] According to the obtained rMxLOX gene sequence, design two primers (SEQ ID NO.3, SEQ ID NO.4), the upstream primer plus Bam H I recognition sequence, downstream primer plus Hind ШRecognition sequence:
[0039] Add components according to the following PCR system to amplify the LOX gene:
[0040] The PCR program is: 94°C for 3min; 30×(94°C for 40s; 53°C for 50s; 72°C for 90s); 72°C for 10min.
[0041] Purify the PCR product with Shanghai Sangon PCR Product Purification Kit, add Bam H I. Hind ШDouble restriction enzyme digestion, connection with appropriate amount of vector pET-28a digested with the same restriction enzyme, and transformation into Escherichia coli DH5α. Randomly pick a few colonies from the transformation plate, insert them into LB liquid medium, shake culture, extract plasmids, perform elect...
Embodiment 3
[0043] Method for the production of Myxococcus aureus DK1622 lipoxygenase gene (rMxLOX) by fermentation in Escherichia coli:
[0044] (1) Transform Escherichia coli expression host strain BL21(DE3) containing rMxLOX expression plasmid pET-28a-rMxLOX (product of Novagen, Germany), and spread it on solid plate medium containing kanamycin (50 μg / ml), 37 Cultivate for 12-16 hours at ℃;
[0045] (2) Pick a single colony, insert it into 50ml seed liquid medium containing kanamycin (50μg / ml), and culture overnight at 70-90rpm 30°C;
[0046] (3) Take the seed liquid according to the volume ratio of 1% and add it to 100ml fermentation medium containing kanamycin (50μg / ml), shake and cultivate at 37°C and 180rpm for 2-3 hours until the OD600 is about 0.6;
[0047](4) Adjust the fermentation temperature to 16°C and shake at 180rpm for 16 hours.
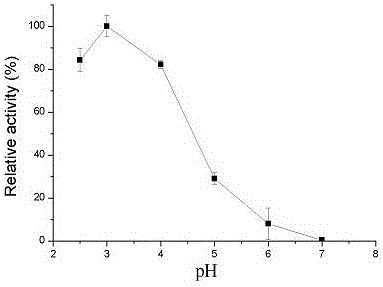
[0048] (5) Collect the bacteria by centrifugation, add pH 3.0 citric acid buffer, break the bacteria by ultrasonic, and collect the supernata...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com