Hydrophobic group modification-based protein N-terminal enrichment method
A hydrophobic group and protein technology, which is applied in the preparation of test samples, material inspection products, biological testing, etc., can solve the problems such as the difficulty of removing materials to eliminate non-specific adsorption, limiting the enrichment selectivity of methods, and the loss of terminal peptides , to achieve high labeling efficiency, promote efficient removal, and high selectivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
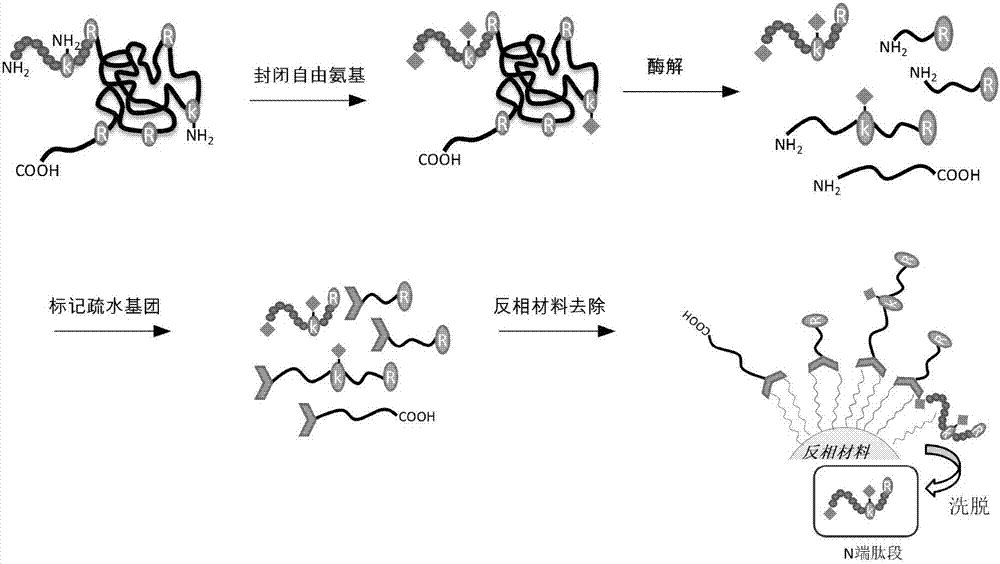
[0030] Such as figure 1 As shown, after the protein is denatured and reductively alkylated, the free amino group is blocked, and then the labeled protein is enzymatically hydrolyzed. The enzymatic hydrolyzed product is labeled with an amino-active reagent with a long carbon chain, and finally the reversed-phase material is used to remove non-N- Telopeptide, thus obtaining the N-terminus of the protein.
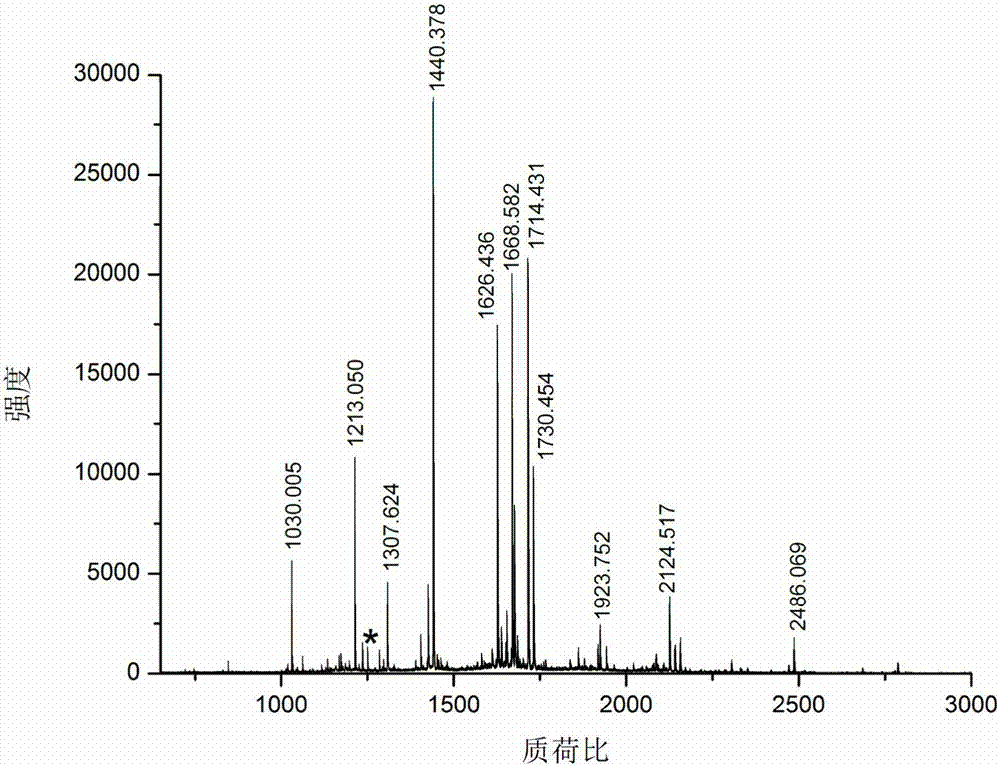
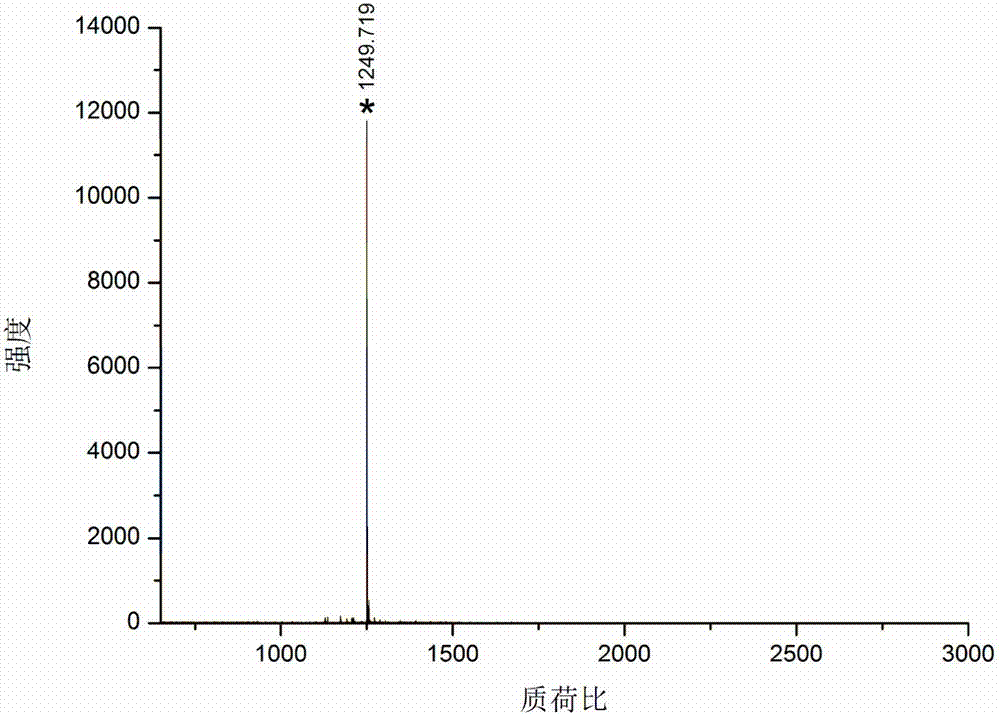
[0031] Take bovine serum albumin and lysozyme as samples, dissolve 100 μg protein in 100 μl 100 mM triethylenediamine carbonate buffer at pH 8 containing 6M guanidine hydrochloride, add 2 μl 100 mM dithiothreitol, and denature at 56 ° C for 1 hour , add 2 μl 300mM acrylamide to react for 1 hour, add another 2 μl 100mM dithiothreitol, incubate for 10 minutes, add final concentration of 100mM formaldehyde and sodium cyanoborohydride to dimethylate the free amino group of the blocked protein, and incubate at room temperature for 2 hours, Transfer the solution to a 10 000 Da ultr...
Embodiment 2
[0034] Take yeast cells as samples, dissolve 100 μg of extracted protein in 100 μl of 100 mM phosphate buffer at pH 8 containing 6M guanidine hydrochloride, add 2 μl of 100 mM dithiothreitol, denature at 56°C for 1 hour, add 2 μl of 300 mM acrylamide for 1 hour Afterwards, another 2 μl of 100 mM dithiothreitol was added, and after incubation for 10 min, a final concentration of 100 mM formaldehyde and sodium cyanoborohydride were added to dimethylate the free amino groups of the blocked protein. After incubation at room temperature for 2 h, the solution was transferred to a 10 000 Da ultrafiltration On the membrane, after removing the solvent by centrifugation and washing residual reagents with 50mM pH 8 4-hydroxyethylpiperazineethanesulfonic acid buffer, the protein was dissolved in 200μl 50mM pH 8 4-hydroxyethylpiperazineethanesulfonic acid buffer middle. Trypsin was added to carry out enzymatic hydrolysis on the ultrafiltration membrane, wherein the amount of enzyme was 1 / 5...
Embodiment 3
[0036] Take human serum as a sample, dissolve 100μg protein in 100μl 100mM phosphate buffer with pH 8 containing 6M guanidine hydrochloride, add 2μl 100mM dithiothreitol, denature and reduce at 56°C for 1h, add 2μl 300mM acrylamide for 1h reaction , add 2 μl of 100mM dithiothreitol, incubate for 10min, add final concentration of 100mM acetic anhydride to block the free amino group of protein, incubate at room temperature for 2h, transfer the solution to a 10 000Da ultrafiltration membrane, centrifuge to remove the solvent and use 50mMpH 8 After washing residual reagents with 4-hydroxyethylpiperazineethanesulfonic acid buffer, the protein was dissolved in 200 μl of 50 mM pH 8 4-hydroxyethylpiperazineethanesulfonic acid buffer. Trypsin was added to carry out enzymatic hydrolysis on the ultrafiltration membrane, wherein the amount of enzyme was 1 / 50 (w / w) of the sample mass, the temperature was 37°C, and the enzymatic hydrolysis was performed for 12 hours. The ultrafiltration mem...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com