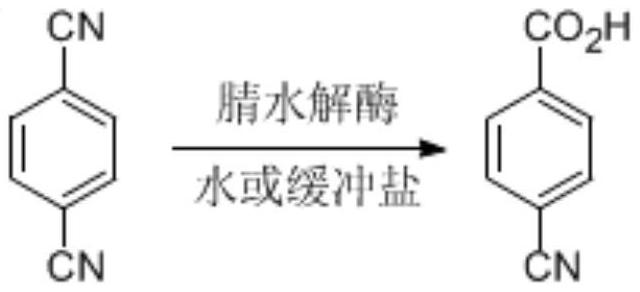
A nitrilase that can hydrolyze terephthalonitrile to p-cyanobenzoic acid
A technology of terephthalonitrile and cyanobenzoic acid, which is applied in the fields of hydrolytic enzymes, genetic engineering, and plant genetic improvement, and can solve problems such as low substrate concentration, low enzyme catalytic efficiency, and long reaction time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Embodiment 1: the acquisition of highly expressed genetically engineered bacteria
[0024] The whole gene synthesis was completed by Shanghai Xuguan Company.
[0025] According to the gene NIT1 (WP_021186296.1) of Pantoea sp.AS-PWVM4, the gene NIT2 (NP_190016) of Arabidopsis thaliana, and the gene NIT3 (ABD98457.1) of Acidovorax facilis72W ), the gene NIT4(U9VXK5) of Leptolyngbya sp., the gene NIT5(XP_013627177.1) of Brassica oleracea var. oleracea and the gene NIT6( XP_010514769.1) were codon-optimized in order to enable the gene to be expressed in the E. coli expression host, and the sequence is shown in the attached table. And add corresponding enzyme cutting sites at both ends of the gene, and construct them into corresponding vectors to obtain genetically engineered bacteria N1, N2, N3, N4, N5, N6.
[0026] Transform the prepared recombinant vector into Escherichia coli BL21, Rosetta or Origami by conventional methods to construct a genetically engineered bacteri...
Embodiment 2
[0027] The cultivation of embodiment 2 genetically engineered bacteria and the preparation of resting cells
[0028] Pick a single colony on the plate and inoculate it into 5ml of fermentation medium containing corresponding antibiotics, cultivate it for about 15 hours as a seed solution, inoculate it into 600ml of fermentation medium according to the inoculation amount of 1%, and cultivate it on a shaker at 37°C and 200rpm to OD 600 = about 0.6-0.8, add IPTG with a final concentration of 0.1 mM to induce for more than 10 h, and collect the bacteria by centrifuging the culture solution at 8000 rpm.
Embodiment 3
[0029] Embodiment 3 Utilizes resting cell of N1 to catalyze the single hydrolysis of terephthalonitrile
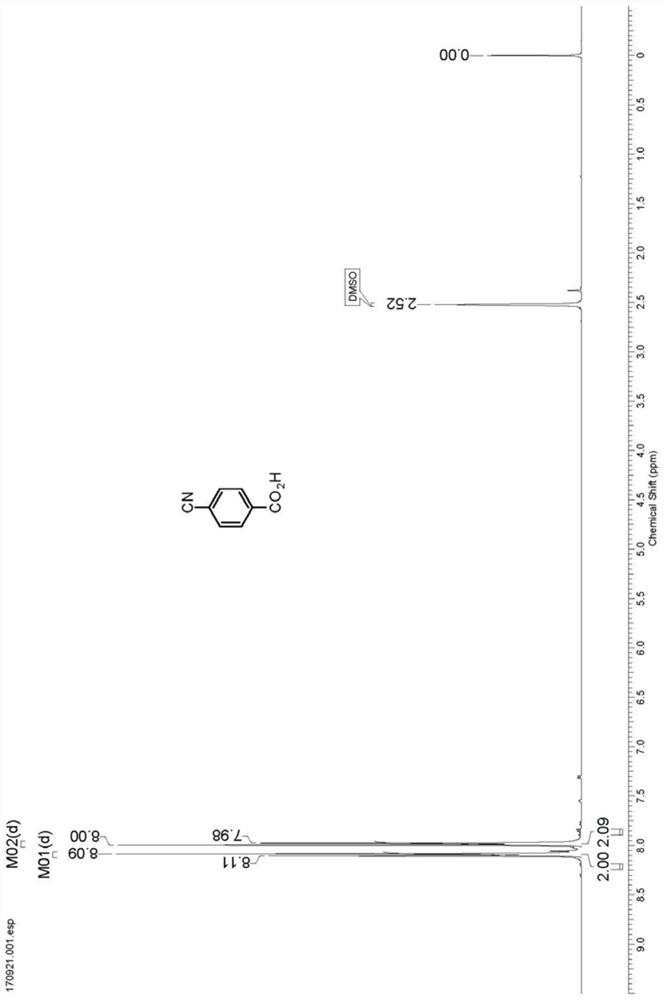
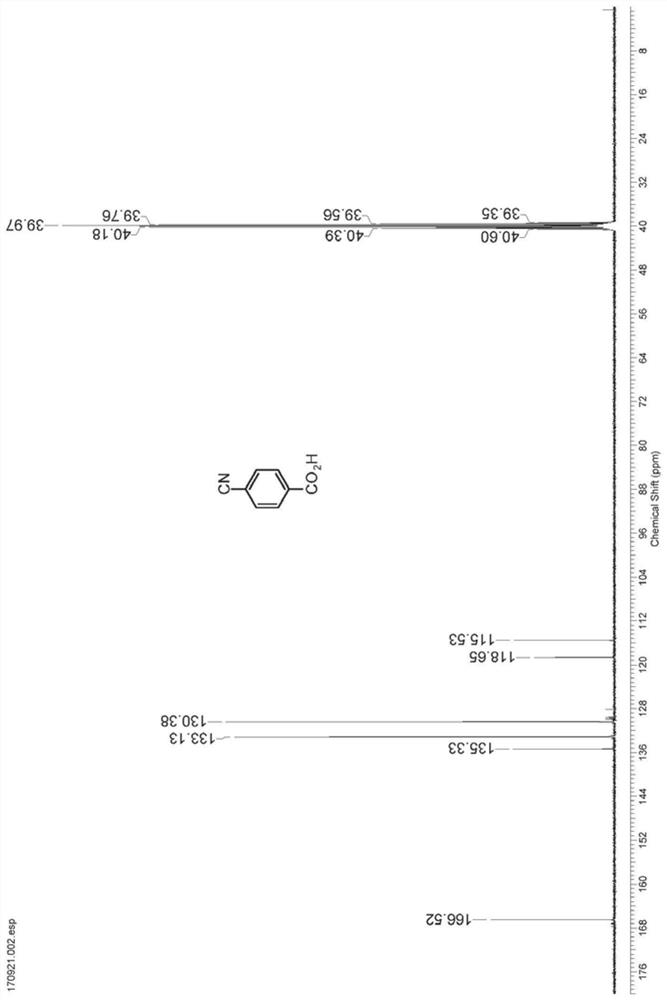
[0030] Take 2.0g N1 resting cells and resuspend in 90mL sodium phosphate buffer (100mM, pH 7.2), add 10mL dimethyl sulfoxide in terephthalonitrile (10.0g) suspension, and then at 30℃, 200rpm The shaking table reacted for 6 hours, and HPLC detection showed that the reaction was complete (liquid chromatography column: Agilent Eclipse XDB-C18 5 μm, 4.6 × 150mm, mobile phase: water (0.5% trifluoroacetic acid) / methanol=75:25, detection wavelength: 230nm, flow rate: 1.0mL / min), centrifuge to recover resting cells, collect the supernatant, acidify to pH 2 with 6M hydrochloric acid, filter with suction, and recrystallize from ethanol to obtain 10.28g of p-cyanobenzoic acid with a yield of 90%.1 H NMR (400MHz, DMSO-d 6 ): δ8.10 (d, J=8.3Hz, 2H), 7.99 (d, J=8.3Hz, 2H). 13 C NMR (100MHz, DMSO-d 6 ): δ166.52, 135.33, 133.13, 130.38, 118.65, 115.53.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com