Engineered yeast strain for expressing pseudomonas putida creatininase and application thereof
A technology of Pseudomonas putida and yeast engineering, applied in microorganism-based methods, enzymes, enzymes, etc., can solve the problems of large demand for creatinine, difficult cultivation of wild bacteria, low specific enzyme activity, etc., to achieve temperature stability and pH. Good stability, easy control of induction conditions, fast and simple cultivation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
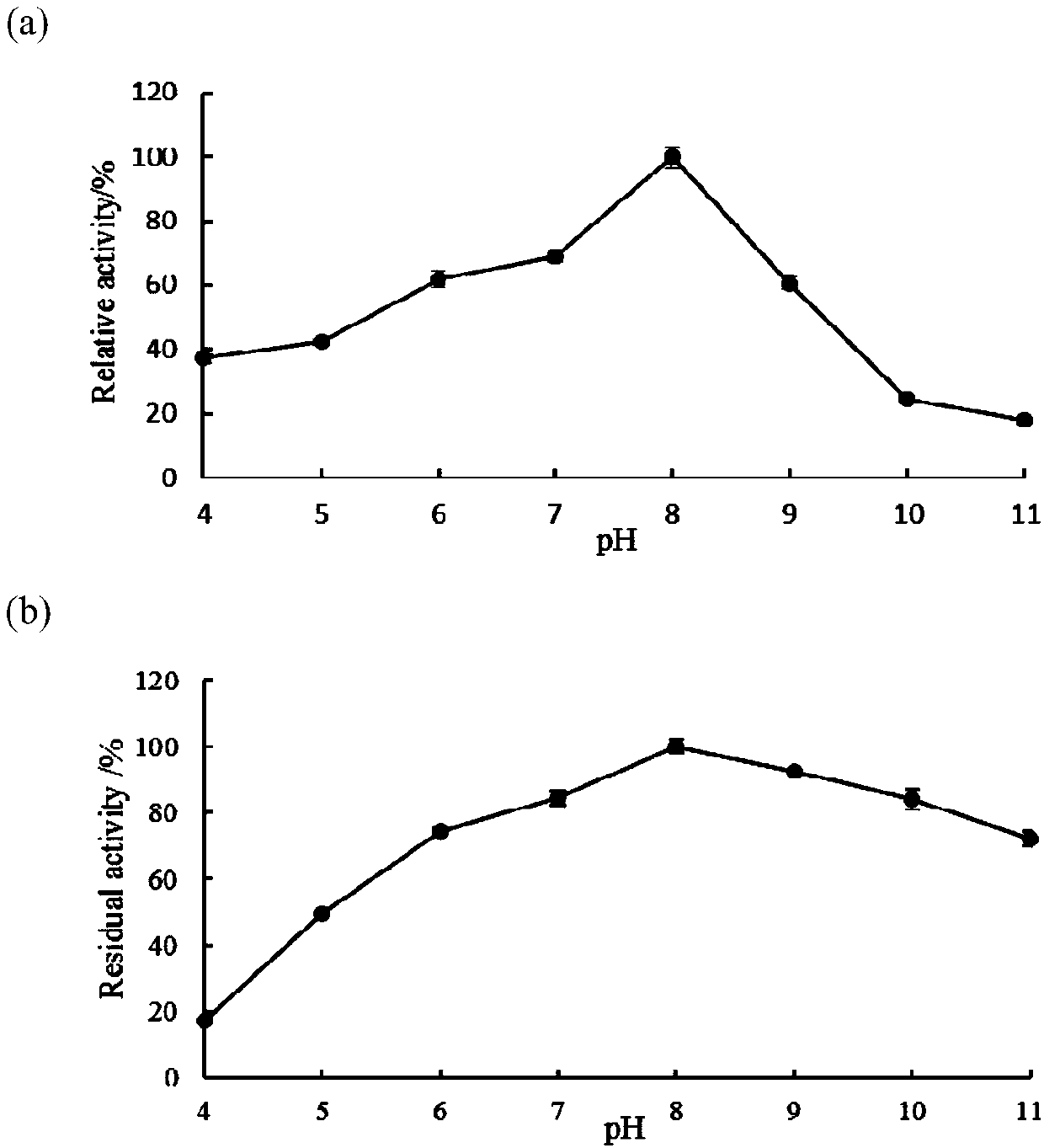
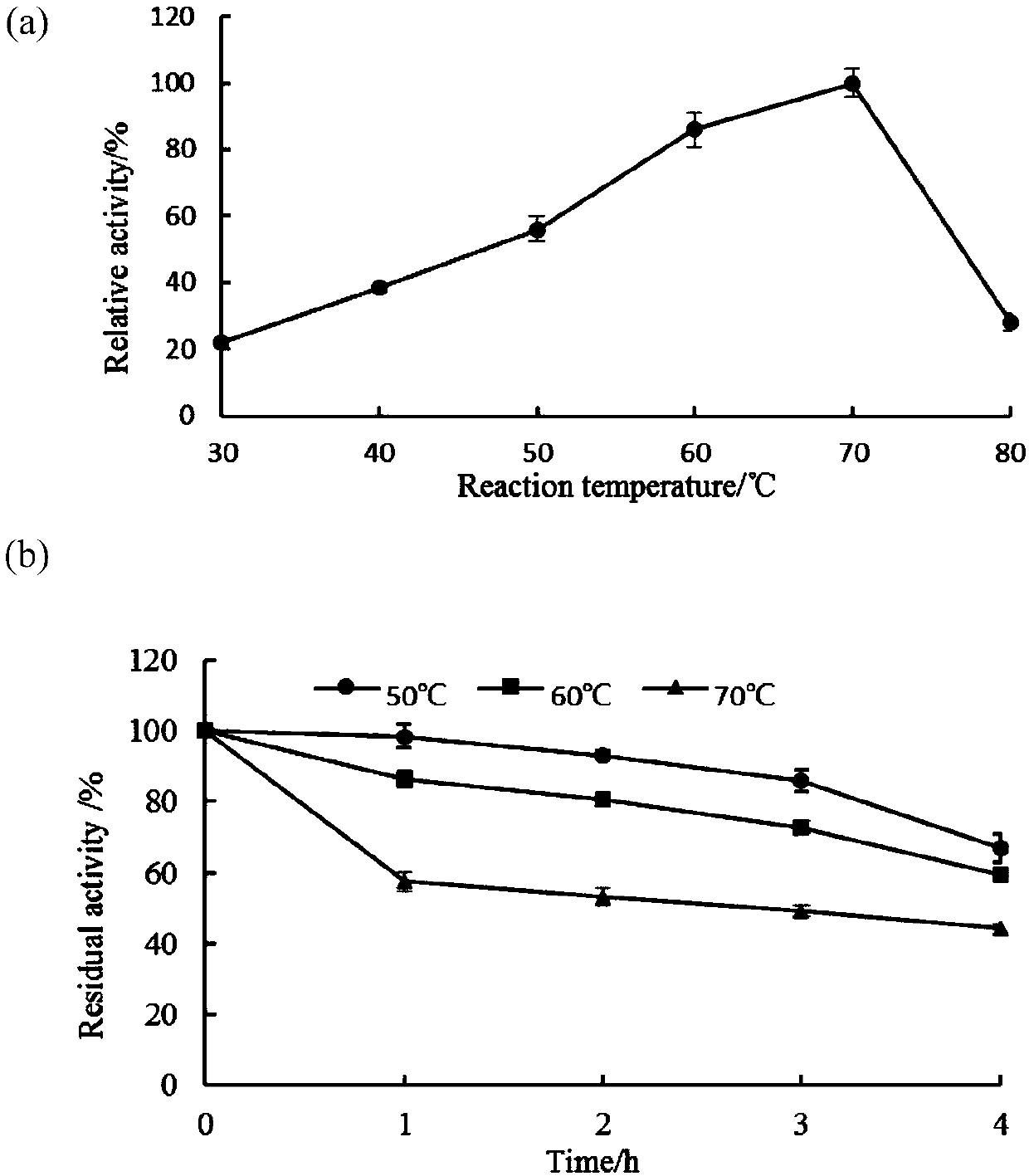
Image
Examples
Embodiment 1
[0031] Synthesis of Novel Creatinase Gene
[0032] Based on the genbank GI:34810367 gene sequence published on the National Center for Biotechnology Information (NCBI, http: / / www.ncbi.nlm.nih.gov / ), the sequence was carried out according to the codon preference of Pichia pastoris Optimized to obtain a novel creatinase gene, its sequence is shown in SEQ ID NO.1, through biotechnology company (Shanghai Jierui Bioengineering Co., Ltd.) whole gene synthesis, the synthetic gene clone in pGH plasmid (purchased from Jierui Bioengineering Co., Ltd. Co., Ltd.) to obtain the plasmid pGH-cre.
Embodiment 2
[0034] Construction of Recombinant Plasmid pHKFA1-cre Containing Novel Creatinase Gene and E.coliTop10 / pHKFA1-cre Carrying the Plasmid
[0035] Design PCR primers according to the nucleotide sequence of the novel creatinase gene: Forward primer cre-F: 5'-CGGGAATTCATGCATCATCATCATCATCATTCTAAGTCTGT
[0036] TTTT-3' (the underline is the EcoRI restriction site, which also contains a 6×His tag) and the reverse primer cre-R: 5'-GATGCGGCCGCTTAAGTTGGTGGGAAC-3' (the underline is the NotI restriction site, which also contains a stop codon) .
[0037] The plasmid pGH-cre in Example 1 was used as a template, and cre-F and cre-R were used as primers for PCR amplification. The gel-recovered and purified PCR product was double-digested with restriction endonucleases EcoRI and NotI, and ligated with the plasmid pHKFA1 that had also been double-digested with EcoRI and NotI with T4 ligase overnight at 16°C, and the ligated product was chemically transformed into E.coliTop10 (purchased from In...
Embodiment 3
[0039] Construction of a Novel Creatinase-producing Strain PichiapastorisGS115 / pHKFA1-cre and Expression of Recombinant Creatinase
[0040] The recombinant plasmid pHKFA1-cre was linearized and purified by Kpn2I single enzyme digestion, and then electrotransformed into Pichiapastoris GS115 (purchased from Invitrogen Life Technologies Co., Ltd., USA) competent cells, and positive transformants were screened on MD plates. The transformants screened on the MD plate were used as templates, and cre-F and cre-R were used as primers for PCR identification. The correct transformants identified by PCR were preliminarily identified as the new creatinase-producing strain PichiapastorisGS115 / pHKFA1-cre.
[0041]Pick 3 transformants identified by PCR and inoculate them into BMGY medium, culture at 30°C, 250rpm until OD600 is close to 6.0, collect cells by centrifuging at 6000rpm, 4°C for 5min, and then resuspend the collected cells in BMMY medium to start The OD600 is close to 1.0, culture...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com