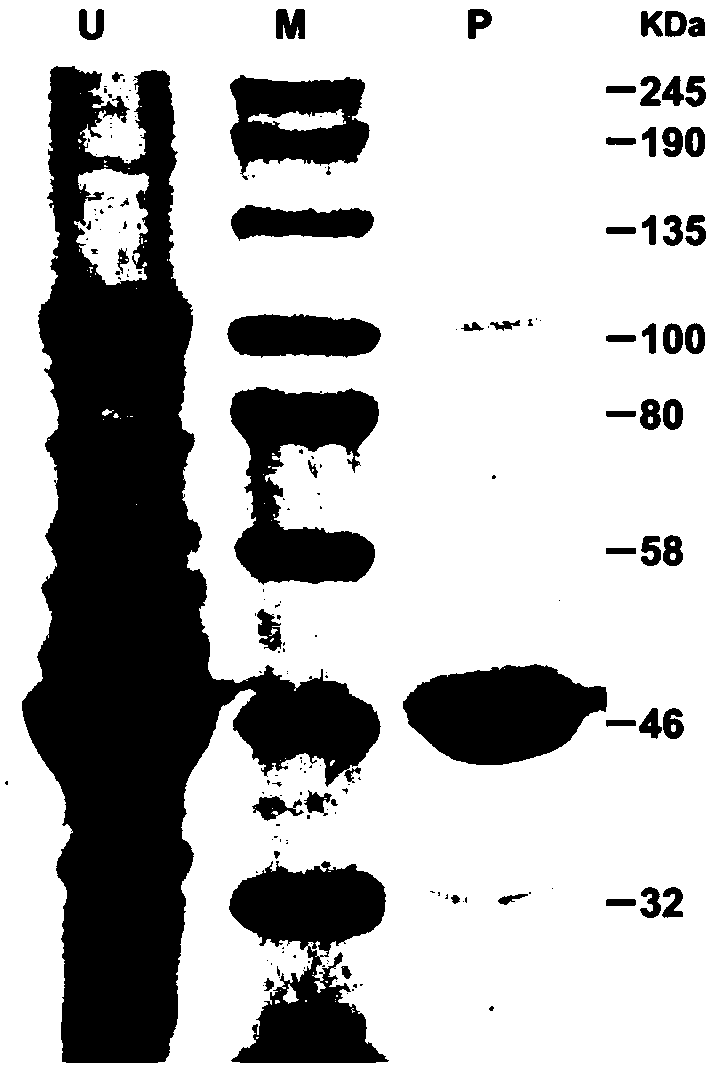O-methyltransferase participating in citrus peel flavone synthesis and coding gene and application thereof
An oxygen methyltransferase, citrus peel technology, applied in transferase, application, genetic engineering and other directions, to achieve high application value and research prospects, the effect of simple enzymatic reaction conditions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Example 1: Cloning of the CitOMT1 gene
[0026] (1) Experimental method
[0027] Gene cloning: CitOMT1 was obtained by performing BLAST analysis on the reported OMT gene of Arabidopsis thaliana and the citrus genome database (http: / / www.citrusgenomedb.org / ), the nucleotide sequence of which is SEQ: No.2. Combining the primer pair SEQ:No.3 and SEQ:No.4, using the cDNA of the oil cell layer of Oumandarin fruit as a template, and referring to the FastStart High FidelityPCR System (Roche) for PCR amplification, the PCR system is: 10×Buffer 3μL, dNTP 2.4 μL, 1.2 μL each of upstream and downstream primers (10 μM), 0.3 μL enzyme, 21.3 μL DEPC-treated water, 0.6 μL cDNA; PCR program: 95 °C pre-denaturation for 5 min; 95 °C denaturation for 30 sec, 58 °C annealing for 30 sec, 72 °C Extend 1min, 35 thermal cycles; 72 extend 1min, store at 4°C. The PCR product was recovered by 1% agarose gel electrophoresis and connected to pGEM-T Easy vector (Promega) for sequencing analysis. ...
Embodiment 2
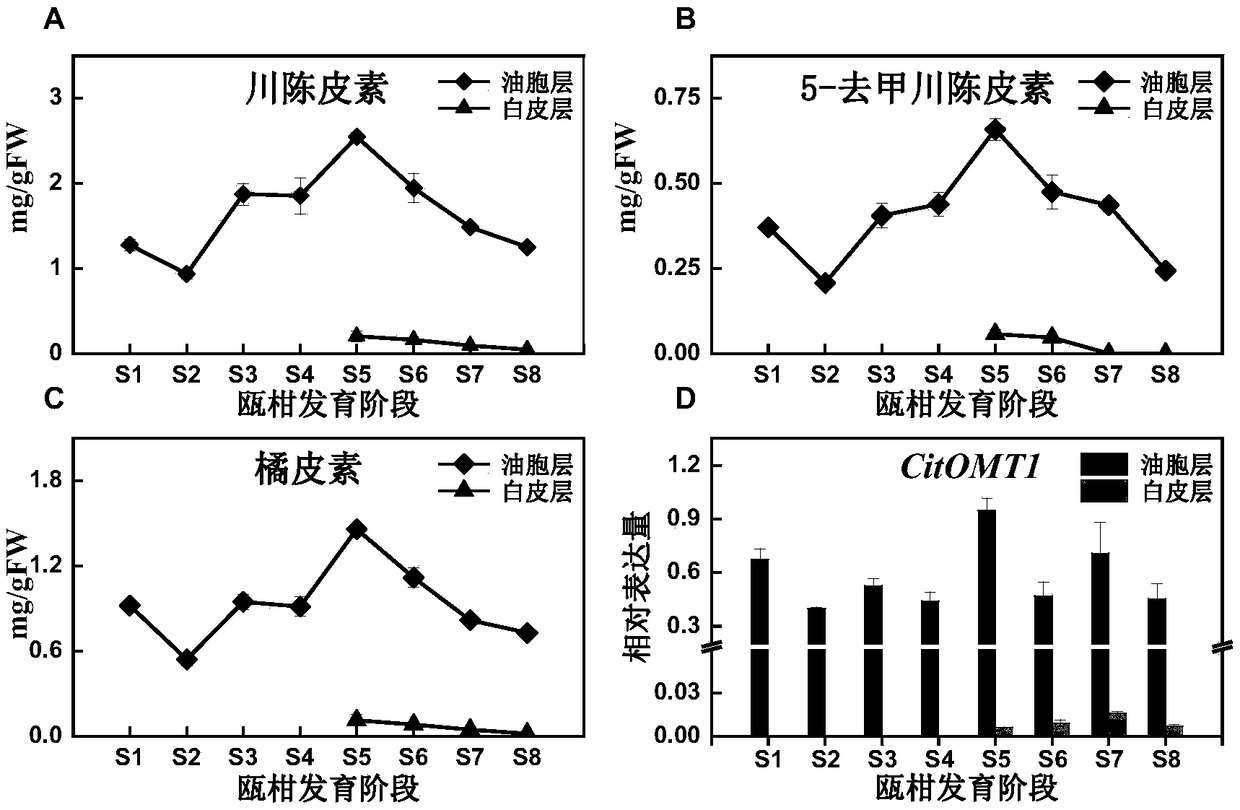
[0030] Example 2: Analysis of the expression pattern of CitOMT1 in the developmental stage of Ou mandarin
[0031] (1) Experimental method
[0032] 1. Fruit material collection
[0033] Ou mandarin fruit of different developmental stages was used as material. Picked at 30(S1), 60(S2), 80(S3), 100(S4), 120(S5), 140(S6), 170(S7), 200(S8) days after full flowering, divided into 3 Biological replicates, 8 fruits were picked for each replicate; the peel of Ou mandarin was divided into two parts, the oil cell layer and the white cortex, cut into small pieces, put them in liquid nitrogen quickly, and then store them at -80°C.
[0034] 2. High performance liquid chromatography (HPLC) analysis of flavonoids
[0035] Weigh 0.5g of the sample powder of Ou mandarin pericarp oil cell layer and white cortex layer, sonicate with 3mL of 80% ethanol for 30min, centrifuge the extract (4000rpm, 10min), repeat three times, combine the supernatant (about 9mL), and use it for the flavonoid group...
Embodiment 3
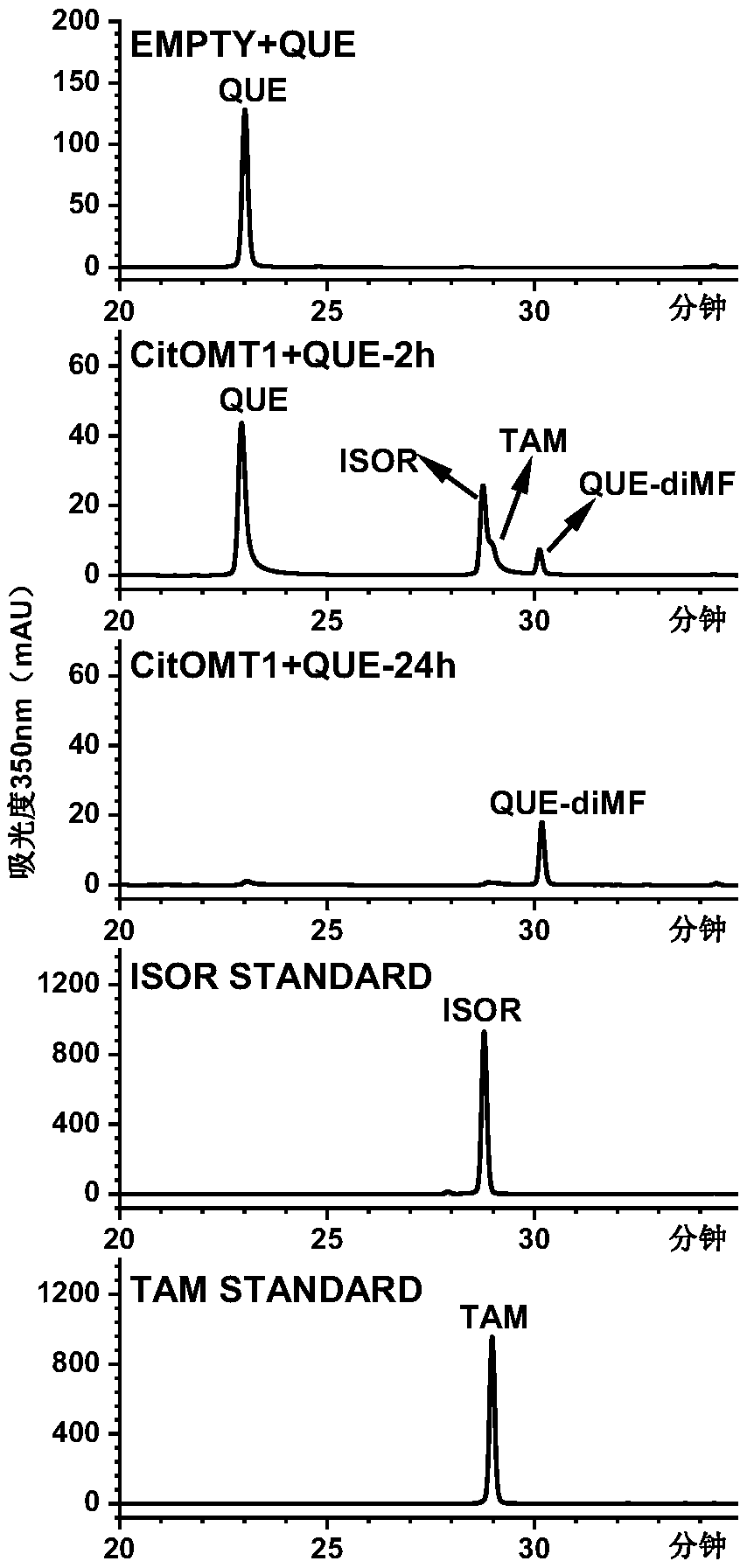
[0044] Example 3: Identification of CitOMT1 Enzyme Activity in Vitro
[0045] (1) Experimental method
[0046] 1. Recombinant vector and recombinant cell construction
[0047] The verified primer pair SEQ:No.3 and SEQ:No.4, plus pET32a restriction site BamHI, XhoI and protection base constitute a new primer pair SEQ:No.10 and SEQ:No.11. Using the sequence-verified CitOMT1-T vector as a template, combined with PCR technology to clone the open reading frame of CitOMT1, and put it on the pET32a vector to construct a CitOMT1-pET recombinant expression vector, and introduce it into the E. coli expression strain by heat shock method In BL21(DE3)pLysS (Promega), spread the bacterial solution evenly on LB plates containing 100 μg / mL ampicillin (Amp) to screen positive single colonies, and store them in 20% glycerol at -80°C for later use.
[0048] 2. Induction and purification of CitOMT1 protein
[0049] After inoculating and activating the glycerol bacteria stored at -80°C, pick a...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com