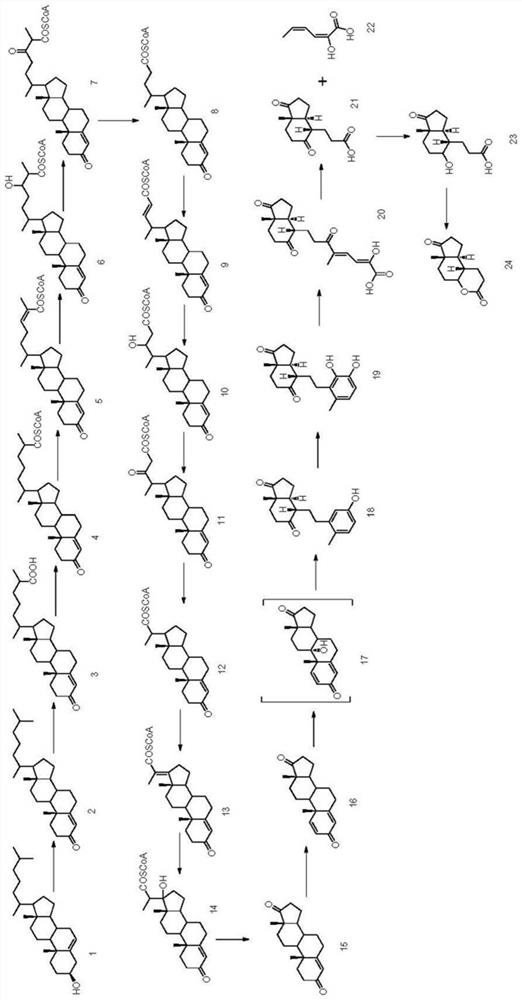
Method, engineering bacteria and application of microbial fermentative production of glutaractone
A microbial fermentation and production engineering technology, applied in the field of microbial fermentation to produce glutaractone, can solve the problems of many by-products, high price of related steroid drugs, high production cost, etc., to block the formation of by-products and improve the conversion rate of substrates , the effect of reducing production costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0081] Embodiment 1: Construction of gene knockout plasmid
[0082] The nucleotide sequence of fatty acid coenzyme A / carboxylate reductase 1 (car1) is shown in SEQ ID NO:1, and the nucleotide sequence of fatty acid coenzyme A / carboxylate reductase 2 (car2) is shown in SEQ ID NO:3 Show. The plasmids for knocking out car1 and the plasmids for knocking out car2 were respectively constructed according to the following methods.
[0083] Car1 and car2 were used as templates for PCR amplification. The PCT system and primers are as follows.
[0084] PCR system:
[0085]
[0086] PCR program: 3 minutes at 98°C; denaturation at 98°C for 10 seconds, annealing at 58°C for 20 seconds, extension at 72°C for 30 seconds, 30 cycles; 10 minutes at 72°C. The primer sequences used are:
[0087] For car1:
[0088] Upstream fragment primers:
[0089] Up-F: 5'TGTTGCCATTGCTGCAGGCATCTCGCACCATCAGC 3' (SEQ ID NO: 5)
[0090] Up-R: 5'AGGTCACTTGGTCGAGCCAGCGCCGCCTGATTCTC 3' (SEQ ID NO: 6)
[00...
Embodiment 2
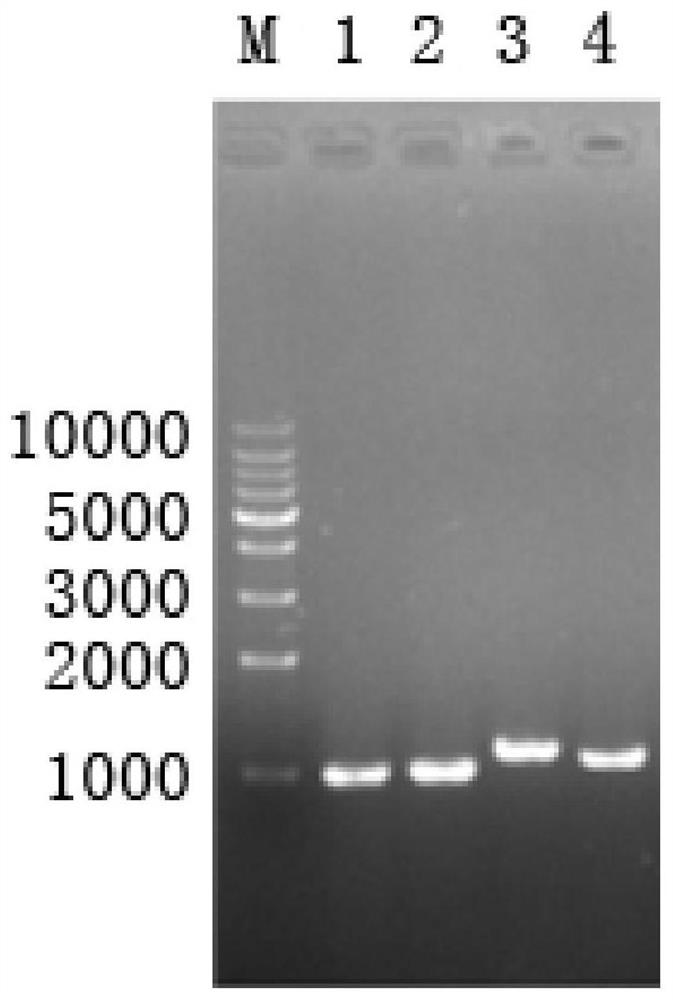
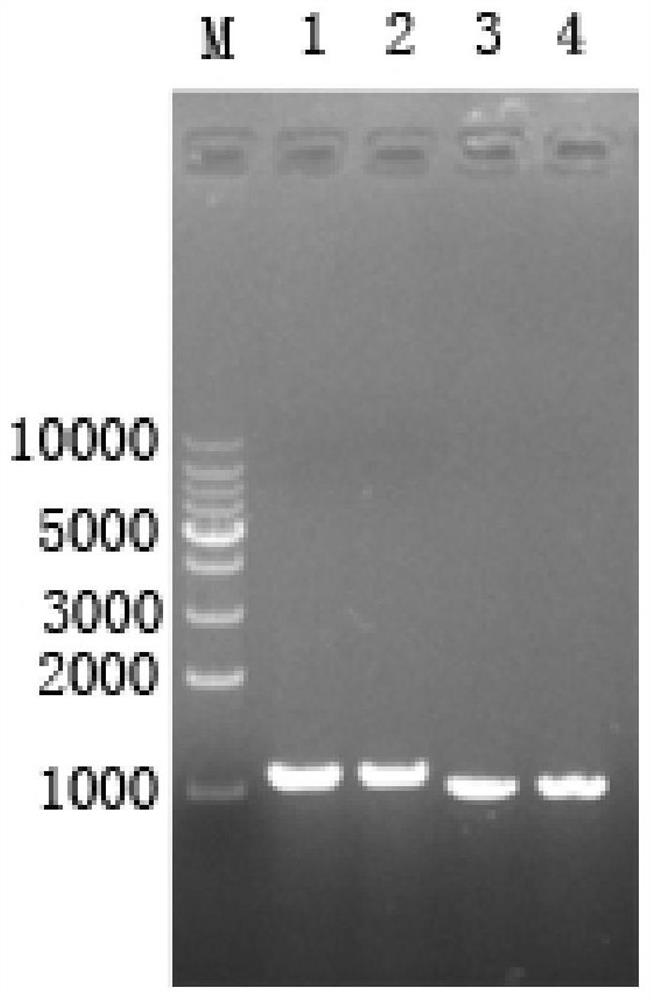
[0102] Example 2: Screening of knockout strains
[0103] The constructed gene knockout plasmid was electrotransformed into Mycobacterium fortuitum (obtained from the American Type Culture Collection (ATCC), strain number: ATCC6841) competent cells, and coated with Kan-resistant LB plates (pancreatic Peptone: 10g / L, yeast extract: 5g / L, sodium chloride: 10g / L, Kan: 50μg / ml, agar: 1.6%) and add IPTG and X-gal for the first screening. Pick blue single colony from it to sucrose plate (tryptone: 10g / L, yeast extract: 5g / L, sucrose: 10g / L, agar: 1.6%) (add IPTG and X-gal), carry out the second time filter. Pick the white colony on the sucrose plate to the liquid LB medium, culture at 30°C for about 36 hours, extract the genome, and use the Up-F and Down-R primers of the target gene for PCR verification. If the gene knockout is successful, the PCR product should be a single fragment of about 2kbp. Figure 5 showed that a mycobacterial strain with car2 gene knockout was successfull...
Embodiment 3
[0104] Embodiment 3: fermentation produces glutaractone
[0105] Seed medium:
[0106] Glucose: 6g / L; Yeast powder: 15g / L; NaNO 3 : 5.4g / L; Glycerin: 2g / L; NH 4 h 2 PO 4 : 0.6g / L; pH: 7.5; sterilized at 115°C for 30 minutes.
[0107] Fermentation medium:
[0108] NaNO 3 : 6.37g / L; KH 2 PO 4 : 1.05g / L; Na 2 HPO 4 : 2.14g / L; MgSO 4 : 0.82g / L; KCl: 0.21g / L; CaCl 2 : 0.1g / L; corn steep liquor dry powder: 14.23g / L; phytosterols: 20g / L; soybean oil: 12%; pH: 7.8; sterilized at 121°C for 30 minutes.
[0109] Fermentation culture steps:
[0110] LB plates (tryptone: 10g / L, yeast extract: 5g / L, sodium chloride: 10g / L, agar: 1.6%) were cultured for 72 hours at 30°C to activate the strains, and activate the bismuth obtained in Example 2 respectively. Knockout strains and wild-type mycobacterial strains (ATCC 6841);
[0111] Inoculate the bacteria from the activated plate into the seed medium, culture at 180 rpm, 30°C for 3 days, and obtain the seed liquid;
[0112] Sampli...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com