Method for improving insect resistance of plants by using RNA interference technique and special DNA fragment of method
A technology of insect resistance and fragmentation, applied in the biological field, can solve the problems of insect resistance evolution, single excellent insect resistance gene, and unsatisfactory insect resistance of transgenic plants, so as to achieve the effect of reducing damage and improving insect resistance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] The gene sequence was obtained by RT-PCR (reverse transcription-polymerase chain reaction) technology.
[0047] 1. PCR primer design
[0048] The PCR primer sequence of chitin deacetylase 5a gene (HaCDA5a) of cotton bollworm is CDA5aF: 5'-ATGAAGTTGTTCGGGCTTCTTG-3', CDA5aR: 5'-CAGTTGTAGATTTATTGTCCAAG-3'.
[0049] 2. Total RNA extraction and cDNA synthesis and amplification of cotton bollworm
[0050] The total RNA of the 5th instar cotton bollworm was extracted with TRIzol reagent, and the first strand of cDNA was synthesized with reverse transcriptase. PCR reaction system: 2 μL 10×Buffer, 1 μmol / L upstream / downstream primers, 0.1 mmol / L dNTPs, 1 unit Taq polymerase, 0.5 μL cDNA, ddH 2 O to make up to 20 μL. PCR reaction program: pre-denaturation at 94°C for 3 min, denaturation at 94°C for 30 s, annealing at 52°C for 30 s, extension at 72°C for 2 min, 35 cycles; final extension at 72°C for 10 min.
[0051] 3. PCR product recovery, TA cloning and identification
[00...
Embodiment 2
[0054] 1. Preparation of target sequence
[0055] PCR technology was used to amplify the above positive clones to prepare target sequences, and the primer sequences were:
[0056] CDAI5aF: 5'-accaggtctcaggagATGAAGTTGTTCGGGCTTCTTG-3',
[0057] CDAI5aR: 5'-accaggtctcatcgtTACTCCAAGCCATATTCCTG-3',
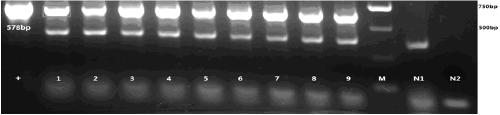
[0058] The lowercase part of the 15 bases at the 5' end of the primer is a partial sequence required by the Golden gate cloning technique. The size of the amplified target gene sequence is 578bp (HaCDA5a, shown as sequence 2 in the sequence listing).
[0059] 2. PCR amplification to obtain target gene fragments
[0060] The PCR amplification system is: 0.6 μL of plasmid template, 2.4 μL of corresponding upstream and downstream primers, 15 μL of 2×Taq MasterMix, ddH 2 Make up 30 μL with O; the PCR amplification program is: 95°C, 3min; 95°C, 30s; 52°C, 30s; 72°C, 40s, 35 cycles; 72°C, 10min. The amplified PCR product was subjected to agarose gel electrophoresis, and the DNA electrop...
Embodiment 3
[0062] Construction of plant hpRNA expression vector
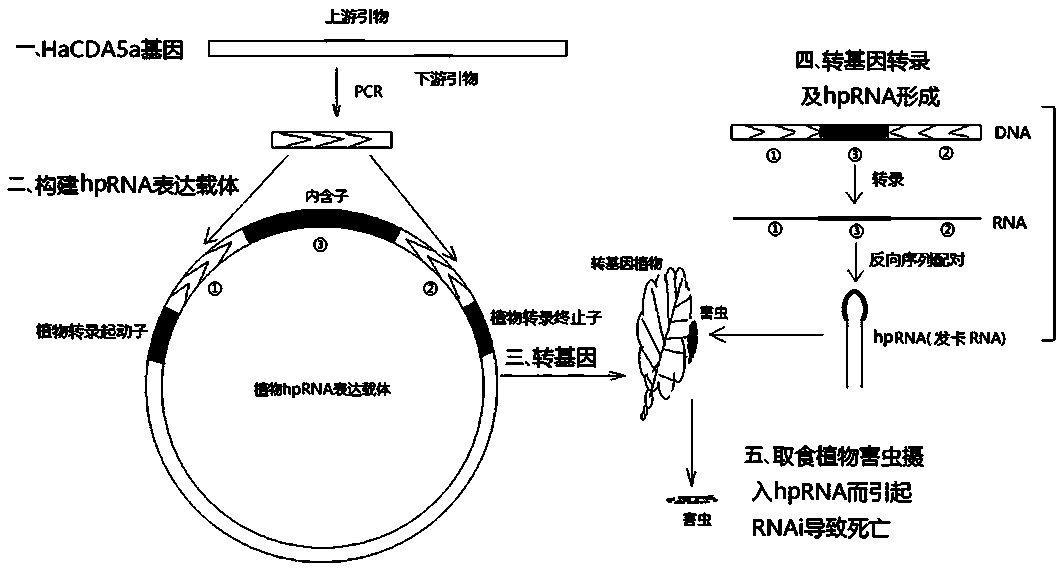
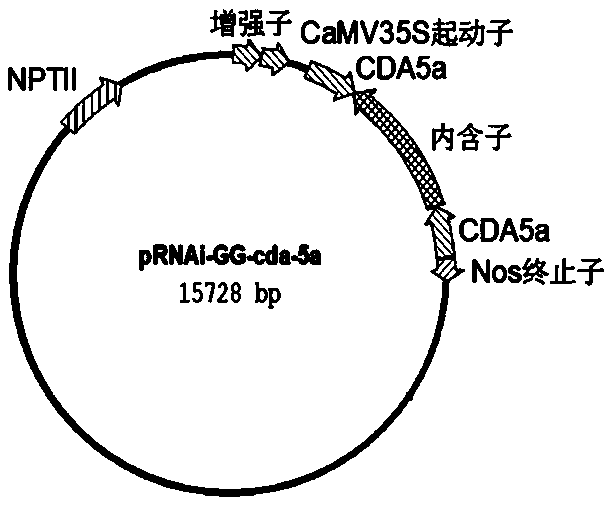
[0063] The backbone of the plant expression vector used in this example is pRNAi-GG, and there is an hpRNA expression cassette on the vector plasmid: driven by the CaMV35S promoter and terminated by the Nos terminator; the transcribed region has two bacterial ccdB separated by an intron sequence Gene; each ccdB gene sequence has two Bsa I endonuclease (IIS type) recognition sites at both ends.
[0064] Plant expression vectors were constructed using Goldgate technology. Put the target sequence PCR amplification product and pRNAi-GG plasmid in one tube, and add Bsa I and T4 DNA ligase (using T4 ligase reaction buffer) at the same time for GoldenGate reaction. The reaction system is: pRNAi-GG plasmid 200 ng , target sequence PCR product 50 ng, T4 ligase 350 U (TAKARA company), T4 ligase buffer 1 μL (TAKARA company), Bsa I 5 U (NEB company), ddH 2 O to make up 10 μL. After transformation, sequencing identification and othe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com