Rapid PCR amplification kit for yeast colonies and method for performing PCR amplification on yeast colonies by using kit
A yeast and kit technology, applied in the field of bioengineering, can solve the problems of genome release PCR amplification failure, cumbersome operation, increase experimental cost and other problems, and achieve the effect of realizing rapid PCR amplification, reducing cross-contamination, and easy to popularize and use.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] A yeast colony rapid PCR amplification kit, including: 10× lysate, 10× PCR reaction solution, dNTP, hot start Taq enzyme;
[0030] The ingredients contained in every liter of 10× lysate are: 20mM KOH, 1.5% (w / v) sodium lauryl sulfate, 60% (v / v) polyethylene glycol;
[0031] The components contained in every liter of 10×PCR reaction solution are: 100mM Tris-HCl, 500mM KCl, 15 mM MgCl2, 0.01% (w / v) gelatin, 0.1% (v / v) polyethylene glycol octyl phenyl ether , pH 8.3 at 25°C.
Embodiment 2
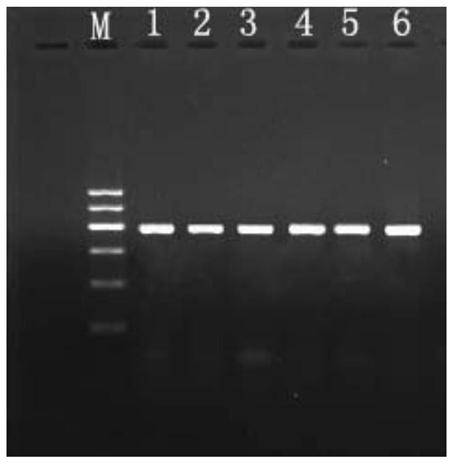
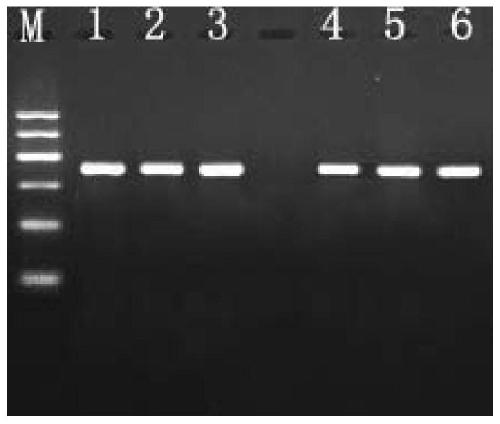
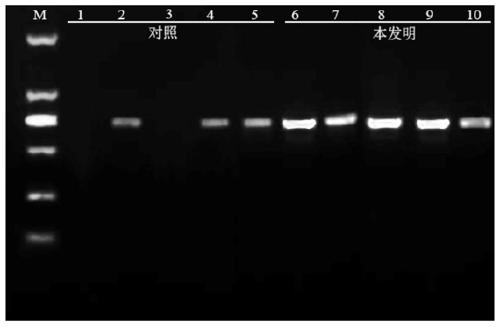
[0033] Utilize the method for PCR amplification Pichia GS115 colony GAP gene of yeast bacterium colony rapid PCR amplification kit in embodiment 1, comprise the following steps:
[0034] (1) Take 500ul of fresh bacterial liquid cultured by Pichia pastoris GS115 monoclonal colony, centrifuge at 13,000 rpm for 20s, discard the supernatant, add 200μL of cell lysate to the centrifuge tube, and use a micro-vortexer to vortex for 15s to make the bacteria and After fully mixing the lysate, let it stand at room temperature for 3 minutes, centrifuge at 13,000 rpm for 30 seconds at 4°C, and use the supernatant for later use.
[0035] (2) Prepare the PCR reaction system including: 2.5 μL of 10×PCR reaction solution, 2.5 mM dNTP, 2.5 μL, 1 μL of 10 umol upstream and downstream primers, 2.5 μL of lysed supernatant, 1 μL of hot-start Taq enzyme, and make up to 25 μL with double distilled water ;
[0036] The upstream primer sequence is: TTCAAGCAGCGTCACTCCTC;
[0037] The downstream primer...
Embodiment 3
[0042] Utilize the yeast bacterium colony rapid PCR amplification kit PCR amplification method in embodiment 1 Pichia GS115 bacterial colony exogenous pYES2-GFP carrier, comprise the following steps:
[0043] (1) Take 500ul of fresh bacterial liquid cultured by Pichia pastoris GS115 monoclonal colony, centrifuge at 13,000 rpm for 20s, discard the supernatant, add 200μL of cell lysate to the centrifuge tube, vortex for 15s to fully mix the bacteria and the lysate Afterwards, let stand at room temperature for 3 minutes, centrifuge at 13,000 rpm for 30 seconds at 4°C, and use the supernatant for later use.
[0044] (2) Prepare the PCR reaction system including: 2.5 μL of 10×PCR reaction solution, 2.5 μL of 2.5mM dNTP, 1 μL of 10 μmol upstream and downstream primers, 2.5 μL of lysed supernatant, 1 μL of hot-start Taq enzyme, supplemented with double distilled water to 25 μL;
[0045] The fragment amplified in this example uses pYES2-GFP universal sequencing primer, the upstream p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com