Application of HMGCR gene in improvement of LV packaging efficiency and infectivity
An infectivity and gene technology, applied in the field of genetic engineering, can solve the problem of not meeting the requirements of cGMP, and achieve the effects of improving titer and infectivity, low cost and simple operation steps.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0073] Example 1 pcDNA4-HMGCR recombinant overexpression vector construction
[0074] This embodiment discloses a method for constructing a pcDNA4-HMGCR recombinant overexpression vector, which specifically includes the following steps:
[0075] 1.1 Synthesis of HMGCR gene sequence
[0076] Look up the gene sequence of NM_000859 in the NCBI database, artificially synthesize the sequence, and design an Apa I restriction site upstream of the sequence, and a Pme I restriction site downstream to obtain the HMGCR gene sequence, and its nucleotide sequence is as shown in SEQ ID NO: 1 shown.
[0077] 1.2 Vector construction
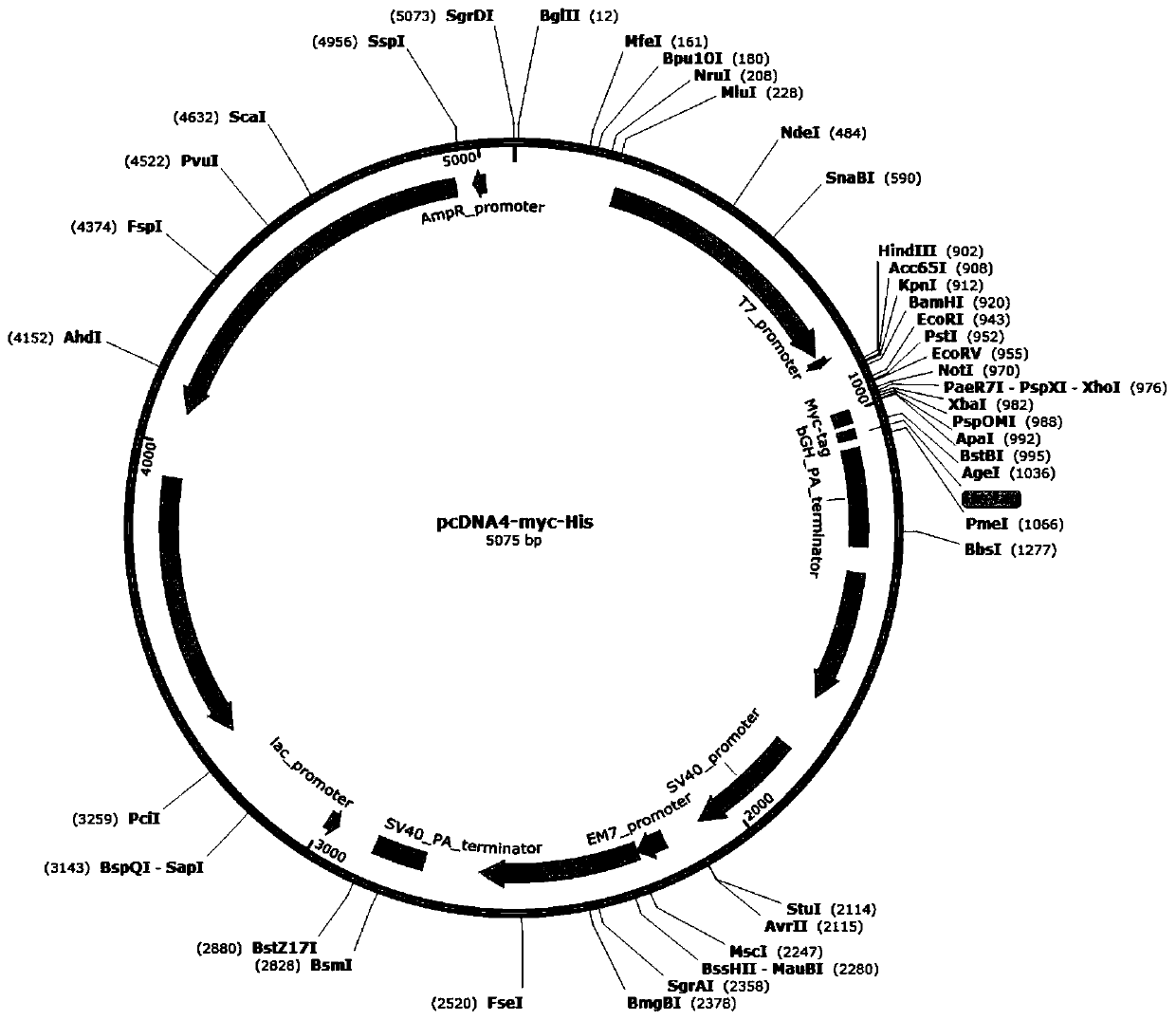
[0078] 1.2.1 Use Apa I and Pme I to digest the pcDNA4-Myc-6His plasmid respectively (the plasmid map is as follows figure 1 shown) and the synthesized HMGCR DNA were subjected to agarose gel electrophoresis after digestion at 37°C for 2 hours. The digestion system was as follows:
[0079] 2μg (2μl)
pcDNA4-Myc-6His or HMGCR DNA
1μl
Apa...
Embodiment 2
[0091] This embodiment provides a method for increasing lentivirus titer, comprising the following steps:
[0092] 1.1 Inoculate 293T cells (purchased from ATCC cell bank in the United States) on cell culture plates.
[0093] 1.2 Carry out the transfection test when the cell confluency reaches 80%; the specific steps are:
[0094] 1.2.1 Preparation of DNA and transfection reagents:
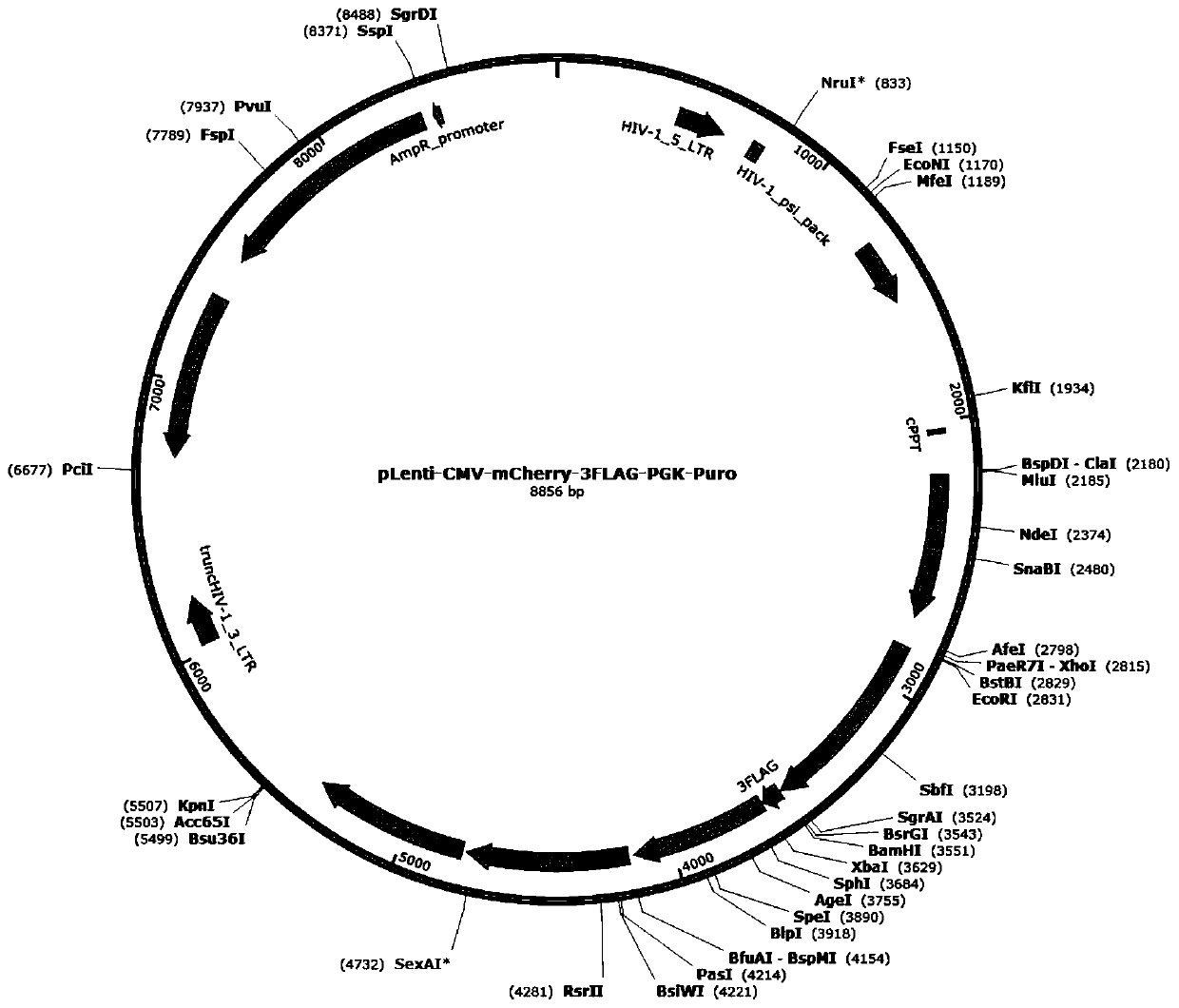
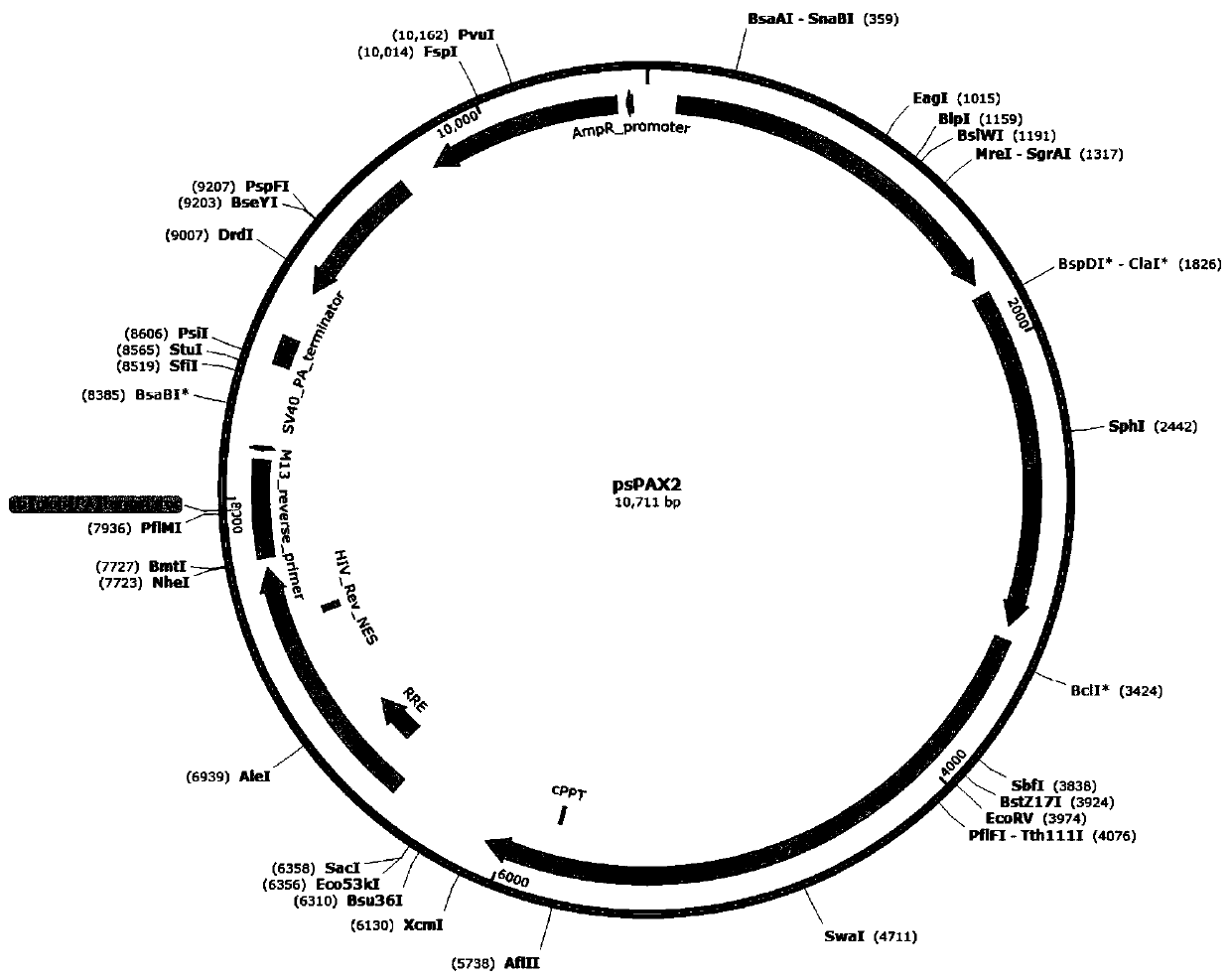
[0095] When the cell culture plate is a 6-well plate, the ratio of each well is plasmid pcDNA4-HMGCR (overexpression well) or PBS (control well): pLenti-CMV-mCherry-3FLAG-PGK-Puro: psPAX2: pHCMV-VSV-G: Transfection reagent (lipofectmine 2000)=lug: lug: lug: lug: 8ul. Among them, the plasmid maps of the lentiviral vector plasmid pLenti-CMV-mCherry-3FLAG-PGK-Puro, the viral packaging auxiliary plasmid psPAX2 and the lentiviral packaging auxiliary plasmid pHCMV-VSV-G are as follows Figure 2-Figure 4 shown.
[0096] 1.2.2 Transfection
[0097] 1.2.2.1 Incubate the diluted DNA and transfection re...
Embodiment 3
[0104] This embodiment provides a method for improving lentivirus infectivity, wherein the steps of the experimental group are as follows:
[0105] S1: co-transfect cells with eukaryotic expression vector and lentiviral packaging system plasmid;
[0106] S2: Collect lentivirus stock solution and concentrate and purify;
[0107] S3: Infect the cells with the concentrated and purified lentivirus stock solution.
[0108] Explanation of terms:
[0109] The MOI value (Multiplicity Of Infection, multiplicity of infection) usually has two meanings: 1) refers to the average number of active units infected with the virus per cell; 2) expresses the number of viruses in terms of virus particles or genome numbers, such as v.p. or v.g., at this time MOI The meaning of refers to the average number of virus particles or genomes per cell infected with the virus.
[0110] The formula for calculating the amount of virus added: (number of cells × MOI value / titer of virus stock solution) × 1...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com