Method for detecting number of soil fungi based on real-time fluorescent quantitative PCR
A real-time fluorescence quantitative and soil technology, applied in the direction of microbial-based methods, microbial measurement/inspection, biochemical equipment and methods, etc., can solve the problem of not being able to obtain reliable information on the number of fungi, not being able to characterize the overall number of soil fungi, and derailing experimental equipment and other problems, to achieve the effect of eliminating the artificial cultivation or leaching extraction steps, accurate and reliable results, and high test sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Embodiment 1: establish the method based on real-time fluorescence quantitative PCR to detect the amount of soil fungus
[0033]Fluorescence designed for the 18S ribosomal rRNA conserved sequence of common soil fungi (Phlebiopsis gigantea, Trichoderma, Heterobasidion parviporum, and Stereum sanguinolentum) Quantitative PCR (qPCR) general primers are as follows:
[0034] Sequence of upstream primer FF390: 5'-CGATAACGAACGAGACCT-3';
[0035] Sequence of downstream primer FR1: 5'-AICCATTCAATCGGTAIT-3', wherein I is inosine base.
[0036] Standard curve template DNA serial dilution concentration: using the known genome base number (40.92Mb) and quality (4.48432×10 -5 ng) and its target-specific fragment copy number (5 / genome), calculate the target-specific fragment copy concentration (copy / μL) in the standard curve template DNA mother solution; take an appropriate amount of mother solution and add sterilized double distilled water according to the dilution formula Adjust ...
Embodiment 2
[0047] The soil used in this example was collected from the southwest foot of Zijin Mountain (32°04′N, 118°50′E) in Nanjing City, Jiangsu Province, and the mixed forest of sweetgum and black pine. Weakly acidic. Remove the litter layer about 1m away from the main trunk of the tree, and use a sterilized 50mL centrifuge tube to take the surface soil (0-10cm). Treatment 1 is the soil of liquidambar pure forest, and treatment 2 is the soil of liquidambar black pine mixed forest. Each treatment is performed 3 times Repeat, use the above method to detect the number of soil fungi.
[0048] The specific operation steps are as follows:
[0049] 1) Pass the soil sample through a 2mm sieve, weigh about 0.3g of fresh soil, use the OMEGA Soil DNA Extraction Kit to extract the total DNA of the soil, dissolve it in sterilized double distilled water, measure the DNA concentration, and place it in a -20°C refrigerator for later use.
[0050] 2) The hyphae of the template Phlebia radiata FBCC...
Embodiment 3
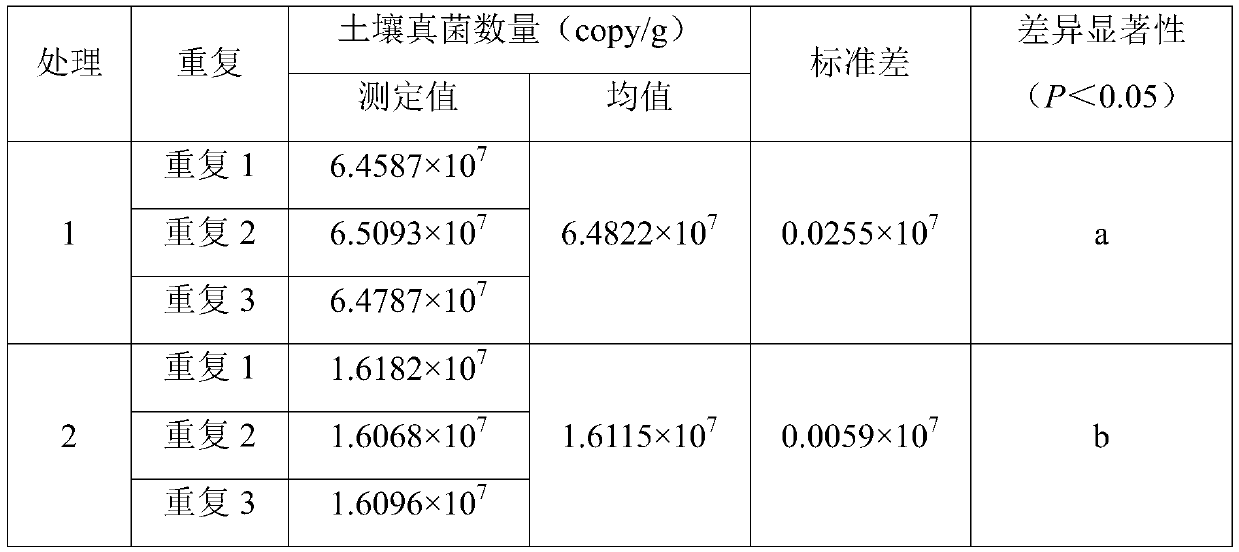
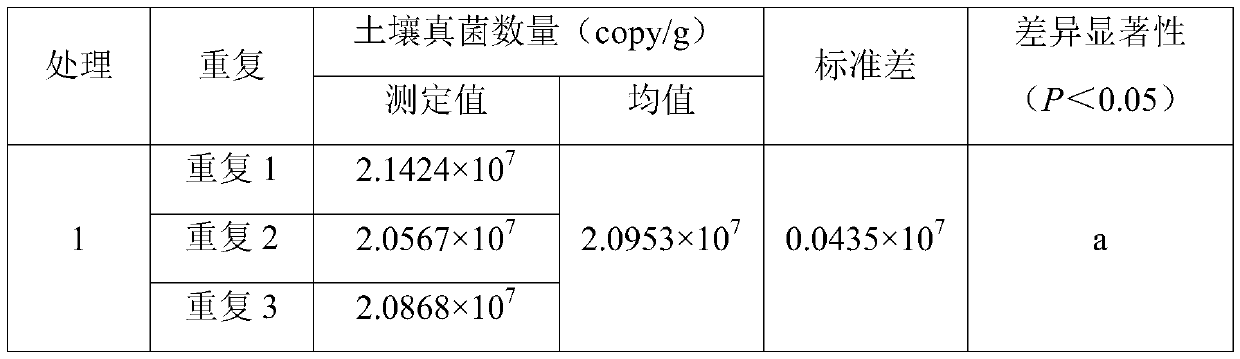
[0062] The soil used in this example was collected from the southern seedling base (21°27'N, 110°11'E) of Zhanjiang City, Guangdong Province, 4-year-old and 9-year-old red eucalyptus plantations. The soil type is brick red soil, which is weakly acidic. Remove the litter layer about 1m away from the main trunk of the tree, and take the surface soil (0-10cm) with a sterilized 50mL centrifuge tube. Treatment 1 is the soil of the 4-year-old red eucalyptus plantation, and treatment 2 is the soil of the 9-year-old red eucalyptus plantation. 3 repetitions. Concrete operation step is with embodiment 2, draws this standard curve, y=-3.4834x+41.119; Wherein R 2 =0.9904, amplification efficiency E=93.68%.
[0063] The test results are as follows:
[0064]
[0065]
[0066] The data in the table also shows the high precision and reproducibility of the results of this analysis method, and also shows that different forest ages have a significant impact on the number of soil fungi.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com