Construction method for human single cell mitochondrial high-throughput sequencing library and kit for library construction
A sequencing library and single-cell technology, applied in the biological field, can solve the problems of undetectable base mutations, high repeatability, lengthy operation, etc., achieve a high rate of false positive site detection rate, strong clinical applicability, The effect of short operating time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
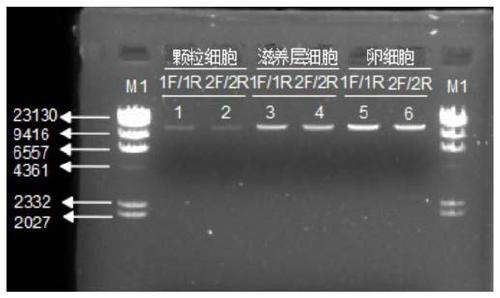
[0059]The following examples are used to illustrate the present invention, but are not intended to limit the scope of the present invention. Unless otherwise specified, the examples are all in accordance with conventional experimental conditions, such as Sambrook et al. Molecular Cloning Experiment Manual (Sambrook J & Russell DW, Molecular Cloning: a Laboratory Manual, 2001), or in accordance with the conditions suggested by the manufacturer's instructions. Example 1 Method for constructing mitochondrial high-throughput sequencing library from a single granulosa cell
[0060] In this example, a single human oocyte peripheral granulosa cell isolated by micromanipulation was used to construct a mitochondrial high-throughput sequencing library.
[0061] The specific operation steps are as follows:
[0062] 1. Single cell collection and lysis
[0063] Collect the patient's granulosa cells, aspirate about 0.5ml (containing hyaluronidase) from the central culture dish, add 1ml of...
Embodiment 2
[0122] Example 2 The method for constructing a mitochondrial high-throughput sequencing library from a single egg cell
[0123] In this example, a single egg cell from a patient isolated by micromanipulation was used to construct a mitochondrial high-throughput sequencing library.
[0124] The specific operation steps are as follows:
[0125] 1. Single cell lysis
[0126] Single human oocytes sorted by micromanipulation were added to 0.2 ml EP tubes pre-packed with 4 microliters of Duchenne's phosphate buffer solution DPBS, and 2.75 microliters of 1 mol / liter dithiothreitol was added with recombination buffer DLB ( Qiagen product number 150343) 0.25 μl, react at 65°C for 10 minutes.
[0127] 2. Separate tube operation of single cell lysate
[0128] Pipette the single-cell lysate repeatedly 10 times with a 10-microliter volumetric pipette, centrifuge briefly, and quickly pipette 3.5 microliters into another 0.2-ml centrifuge tube, divide the cell lysate into two tubes, and m...
Embodiment 3
[0184] Example 3 A single granulosa cell using a linker with a molecular tag (Unique molecular Index, UMI) for mitochondrial high-throughput sequencing library construction
[0185] In this example, a single human oocyte peripheral granulosa cell isolated by micromanipulation was used to construct a mitochondrial high-throughput sequencing library.
[0186] The specific operation steps are as follows:
[0187] 1. Single cell collection and lysis
[0188] Collect the patient's granulosa cells, aspirate about 0.5ml (containing hyaluronidase) from the central culture dish, add 1ml of oviduct fluid culture medium HTF containing 50% SSS to stop digestion, centrifuge at 350g for 1 minute, add HTF containing 10% SSS Centrifuge the culture medium at 350g for 1 minute, resuspend the cell pellet with 1 ml of the above culture medium, take out 10 microliters and add it to the lid of the petri dish, cover with oil, use a microinjection needle with an inner diameter of 20 microliters to s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com