Kits for Escherichia coli and Shigella screening
A technology of Escherichia coli and Shigella, which is applied in the field of protein spectrum detection, can solve the problems of no way to distinguish Shigella populations, high cost, and incorrectness, and achieve no biosafety risk, high accuracy, and low cost Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] Example 1 Determination of the characteristic proteins of Escherichia coli and Shigella
[0055] The invention starts from the biochemical characteristics of microorganisms, investigates the difference in biochemical characteristics of Escherichia coli and Shigella, explores different types of enzymes of Escherichia coli and Shigella, and screens suitable enzyme reactions, aiming to compare the molecular weight of compounds before and after the enzyme reaction, Find the difference between E. coli and Shigella. The types of intracellular / extracellular enzymes of Escherichia coli and Shigella mainly include oxidase, reductase, synthetase, transferase, decarboxylase, dehydrogenase, deaminase, etc. After repeated experiments and multiple verifications, the present invention has determined that the selection principles for identifying the characteristic marker enzymes of Escherichia coli and Shigella are: (1) there is a certain molecular weight difference between the substra...
Embodiment 2
[0059] Example 2 Establishment of the identification method of Escherichia coli and Shigella and the identification of Shiga in Sonnei
[0060] 1. Determination of strain culture method
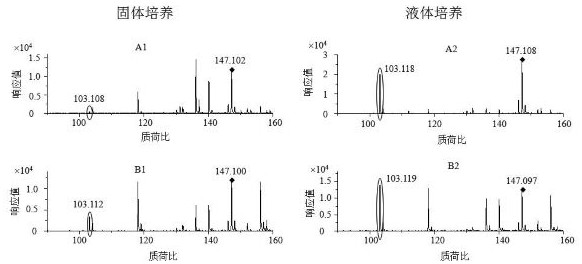
[0061] The invention optimizes and selects the culture conditions before the strain (Escherichia coli or Shigella) enters the reaction solution. Bacterial culture includes solid culture and liquid culture. Escherichia coli was inoculated on LB medium at 37°C for overnight cultivation, scraped and washed with PBS for one time, then inoculated into the reaction solution, incubated at 37°C for 4 hours, centrifuged, and 1µl of the supernatant was spotted on the sample target, and then dried naturally. Overlay with 1 µl of CHCA matrix saturated solution (α-hydro-4-hydroxycinnamic acid saturated in 48.75% acetonitrile, 2.5% trifluoroacetic acid), dried and acquired by MALDI-TOF MS.
[0062] Escherichia coli and Shigella sonnei were respectively inoculated in liquid LB medium and cultured overnigh...
Embodiment 3
[0070] Example 3 Evaluation of the actual detection effect of the kit
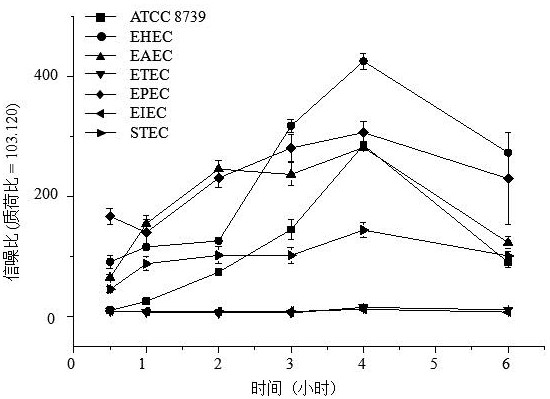
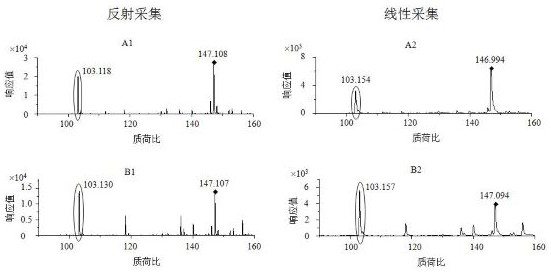
[0071] Using the kit of the invention, 109 strains of bacteria were screened and identified, including 79 strains of Escherichia coli, covering enterohemorrhagic Escherichia coli, pathogenic Escherichia coli, enterotoxigenic Escherichia coli, enteroinvasive Escherichia coli and coagulative large intestine 30 strains of Shigella, including Shigella dysenteriae, Shigella flexneri, Shigella baumannii, and Shigella sonnei. The culture method was liquid culture, the incubation time in the reaction was 2 hours, and the data acquisition mode was the reflection mode. The results are as follows:
[0072]
[0073] The results showed that all 99 strains of Escherichia coli were correctly identified, 12 of the 30 Shigella strains were correctly identified and detected, and the remaining 18 strains were also correctly identified as Shigella, which were detected with traditional biochemical methods. The results wer...
PUM
| Property | Measurement | Unit |
|---|---|---|
| mass-to-charge ratio | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com