Preparation method of peanut mutant, peanut mutant gene, protein encoded by peanut mutant gene and application
A technology for mutant genes and mutants is applied in the field of primers for detecting peanut mutant genes and the preparation of peanut mutants, which can solve problems such as large workload, and achieve the effects of convenient operation, fast mutation speed and high efficiency.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
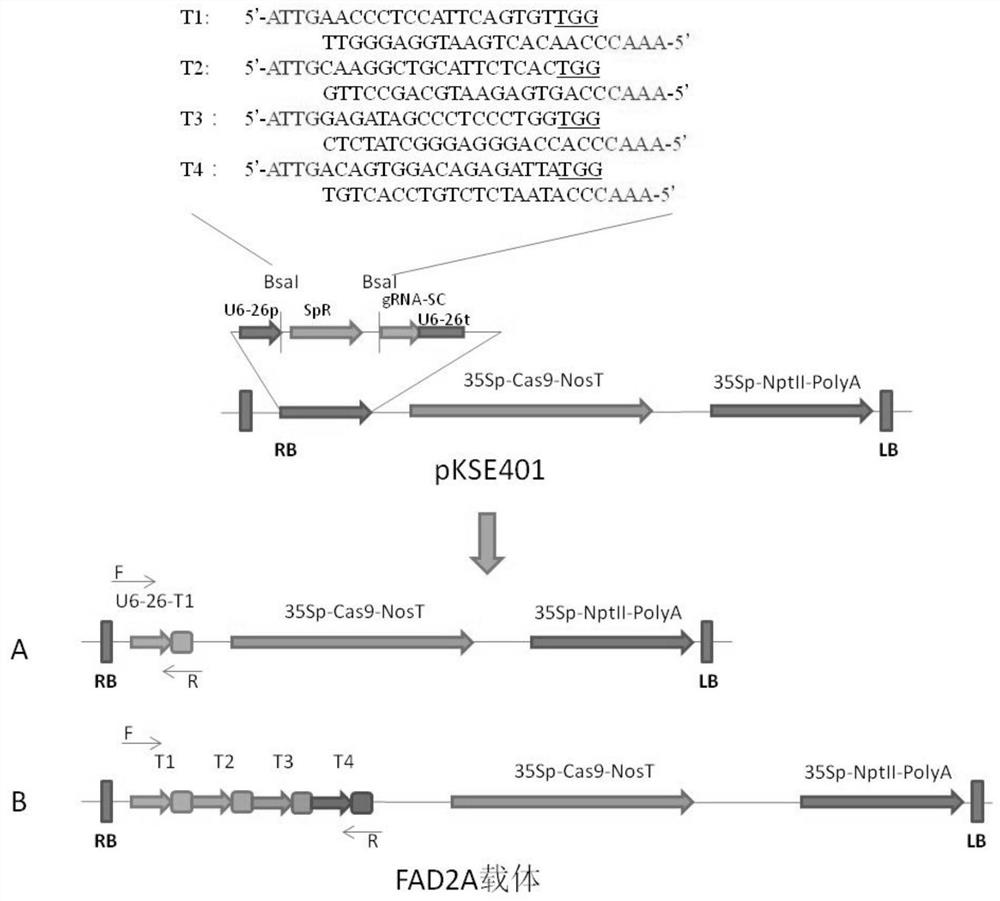
[0035] The embodiment of the present invention provides a kind of preparation method of peanut mutant, it comprises the following steps:
[0036] S1. Provide the coding region sequence of peanut desaturated fatty acidase gene AhFAD2A, CRISPR / Cas9 gene editing vector and peanut embryogenic callus;
[0037] S2. According to the sequence of the coding region of the peanut desaturated fatty acidase gene AhFAD2A, the sgRNA target sequence T1-T4 is obtained;
[0038] S3. According to the sgRNA target sequence T1-T4, synthesize the sgRNA target sequence nucleotide fragments containing cohesive ends and their complementary strands respectively, and mix and anneal to obtain double-stranded DNA T1-T4 containing cohesive ends at both ends;
[0039] S4. Ligate the double-stranded DNA T1-T4 with the CRISPR / Cas9 gene editing vector to obtain the gene editing vector T1-T4;
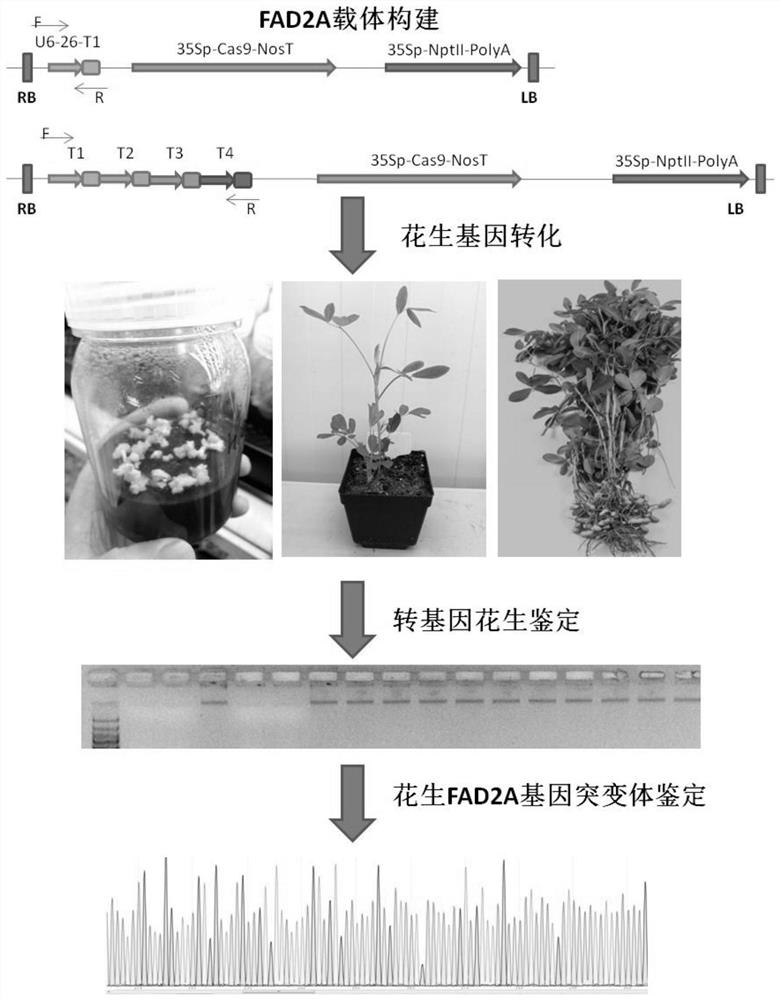
[0040] S5. Transforming peanut embryogenic callus with at least one of the gene editing vectors T1-T4, and culturing ...
Embodiment
[0075] 1. Cloning and sequencing of peanut desaturated fatty acidase gene AhFAD2A
[0076] According to the cultivated peanut (Arachis hypogaea) omega-6desaturase (FadS) mRNA sequence (GenBank: AF030319.1) design PCR primer FAD2A-f and FAD2A-r, its nucleotide sequence is respectively as SEQ ID NO: 6 and SEQ ID NO: 7.
[0077] The peanut leaves were taken, the kit extracted the genomic DNA, and the AhFAD2A gene was amplified by PCR. The PCR reaction system is as follows:
[0078] The 50μl reaction system includes: ddH 2 O: 13 μl, Primer: 5 μl, DNA: 5 μl, 2×Taq mix: 25 μl, Mg 2+ : 1 μl, glycerol: 1 μl.
[0079] The reaction conditions are as follows: pre-denaturation at 95°C for 5 min, followed by a cycle of denaturation at 95°C for 30 s, annealing at 56°C for 30 s, and extension at 72°C for 1 min. After 40 cycles, complete extension at 72°C for 10 min, and storage at 4°C.
[0080] DNA sequencing was performed on the PCR product to obtain the sequence of the coding region o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com