Bacteriophage trp574 gene and application thereof
A phage and gene technology, applied in the phage trp574 gene and its application fields, to achieve the effect of great application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
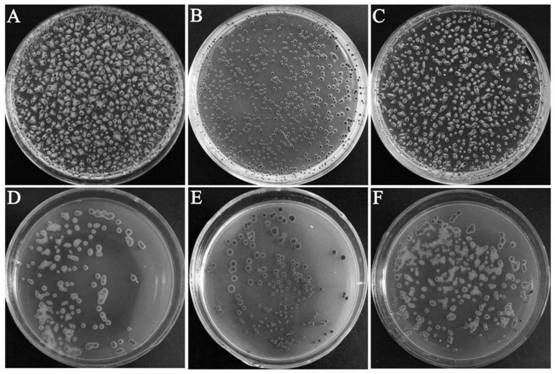
Embodiment 1
[0049] Example 1 Bioinformatics analysis of trp574 gene (orf30)
[0050] In the early stage of the present invention, a transcription regulator orf30 was found by sequencing the genome of the lytic phage P574, and the gene protein sequence was compared using NCBI's Blastp; using the ProtParam tool (https: / / web.expasy.org / protparam / ) to analyze protein physicochemical properties; use SMART server (http: / / smart.embl-heidelberg.de / ) for protein domain analysis; use SWISS-MODEL (https: / / www.swissmodel.expasy.org / ) for Protein model construction.
[0051] NCBI's Blastp results show that the orf30 gene belongs to the XRE family, and the comparison results show that it has 69% homology with the transcription regulator of the Burkholderia phage Bcepmigl belonging to the XRE family. The four protein sequences with the highest scores in the download comparison results are, Use ClustalX software for multiple sequence alignment, and the alignment results are colored by Boxshade (https: / ...
Embodiment 2
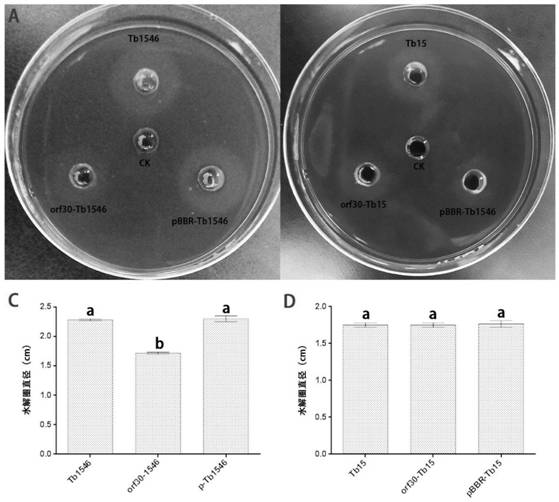
[0052] Embodiment 2trp574 gene (orf30) cloning and recombinant plasmid construction
[0053] 1. Method
[0054] (1) Use the Universal Phage DNA Extraction Kit (Product No.: KG005-1) of Guangzhou Nuojing Biotechnology Co., Ltd. to extract phage DNA, and design the amplification primers orf30-F / orf30-R of the target gene orf30 according to the P574 genome information (such as shown in Table 2); with phage DNA as template and orf30-F / orf30-R as primers, Novozyme MasterMix amplifies the target gene orf30, the amplification system is Master Mix 25μL, Primer1 1μL, Primer2 1μL, template DNA 1μL (2 0 to 50 μL. The PCR amplification conditions were: 95°C for 3min; 95°C for 10s, 55°C for 30s, 72°C for 40s, 35 cycles; 72°C for 10min, 4°C, ∞. After the PCR products were electrophoresed, the target bands in the gel electrophoresis were recovered by gel cutting.
[0055] (2) The pBBR1MCS4 plasmid was extracted according to the instructions of the Axygen plasmid mini-extraction kit, an...
Embodiment 3
[0061] The preparation of embodiment 3 transfer trp574 gene (orf30) solanacearum Tb1546 and Tb15
[0062] 1. Preparation and electrotransformation of R. solanacearum competent cells
[0063] For the preparation of R. solanacearum Tb15 electroshock-competent and the electroshock transformation method, refer to the method of Lavie et al. (2002).
[0064] (1) Ralstonia solanacearum Tb1546 and Tb15 were inoculated in 10 mL NA medium for overnight culture, and the bacterial solution was placed on ice for 15 min, then packed into pre-cooled 1.5 mL centrifuge tubes, centrifuged at 6000 rpm at 4 °C for 5 min, and discarded. supernatant.
[0065](2) Add 1 mL of 10% glycerol to resuspend, centrifuge at 6000 rpm at 4°C for 5 min, and discard the supernatant.
[0066] (3) Repeat step (2) twice.
[0067] (4) Add 100 μL of 10% glycerol to resuspend to obtain Ralstonia solanacearum competence.
[0068] (5) Take 50 μL of prepared Tb1546 and Tb15 Ralstonia solanacearum competently in 0.1 c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com