InDel marker suitable for corn genotyping and application of InDel marker
A genotyping, molecular marker-assisted technology, applied in the fields of genomics, bioinformatics, molecular plant breeding, and molecular biology, it can solve problems such as the inability to guarantee coverage of the target area.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
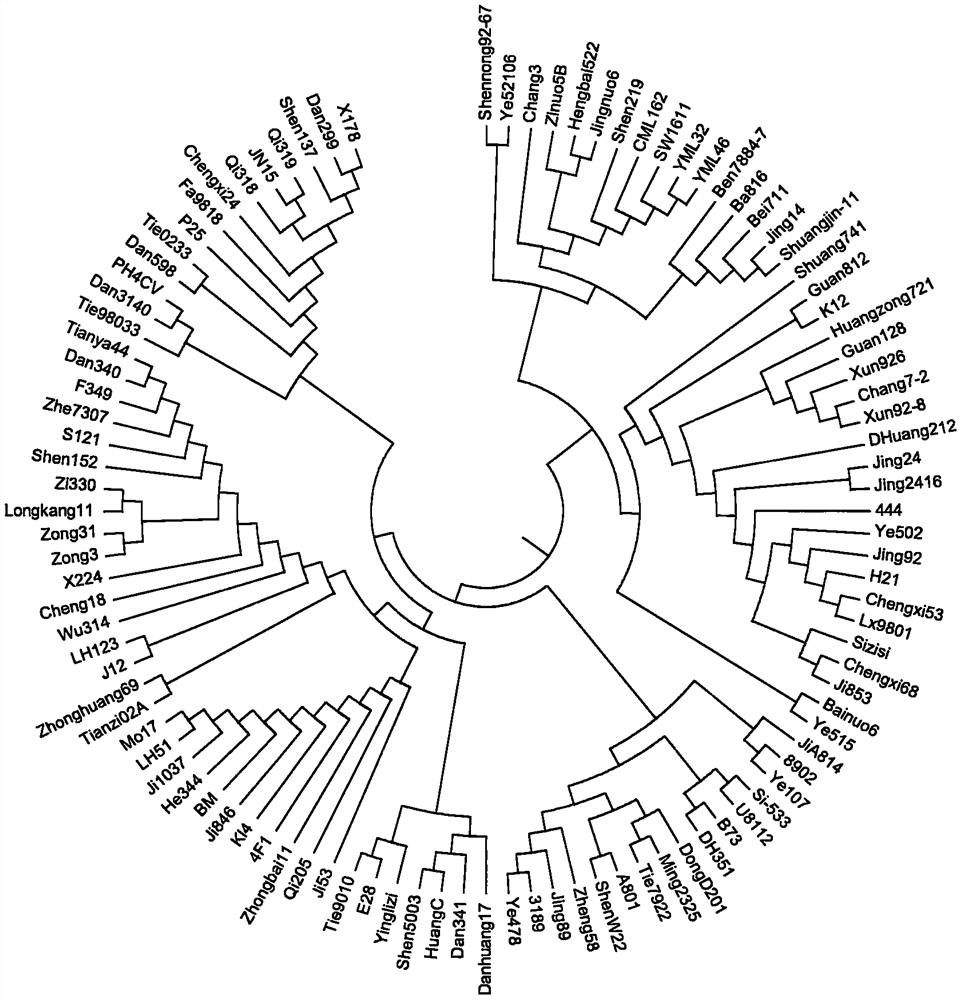
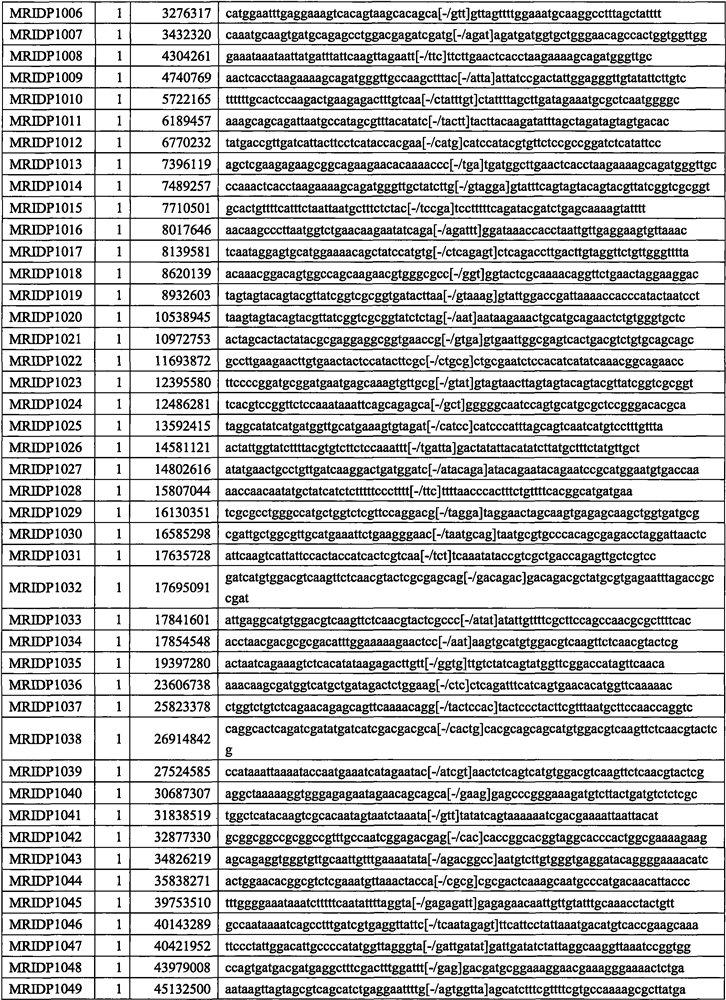
[0024] Example 1 The development process of 1011 InDel markers in corn
[0025] A series of stringent quality control analyzes were performed on the MCIDP40K InDel chip containing 40,793 InDel markers using 500 corn backbone inbred lines and 500 main hybrids in the past 10 years. 24,968 polymorphic high resolution (PolyHigh Resolution, PHR) InDel markers were obtained. These PHR-like InDel markers belong to polymorphic InDel markers that have good genotyping effects on the chip and actually exist on the maize genome. Then the following steps were taken: Step by step screening assessment.
[0026] In the first step, based on the missing data rate of InDel markers, 1218 InDel markers with a missing rate of more than 5% were deleted. After this step, 23750 remaining candidate InDel markers were filtered; in the second step, redundant InDel markers were removed by linkage disequilibrium (LD) Markers, InDel markers belonging to the same linkage disequilibrium segment provide redun...
Embodiment 2
[0053] Example 2 The method of using Ampliseq technology to carry out maize InDel marker genotyping
[0054] (1) Using the CTAB method to extract the genomic DNA of the corn samples to be tested (Jingke 968, Jundan 20, Xianyu 335, Zhengdan 958);
[0055] (2) Construction of multiplex amplification targeted capture sequencing library:
[0056] use The fluorometer quantifies the genomic DNA of the sample to be tested. In the experiment, only 10ng of DNA was needed for each sample, so the DNA solution of the sample to be tested was diluted to 2.5ng / μL as the genomic DNA working solution. The multiplex amplification targeted capture uses the reagent Ion AmpliSeqTM Library Kit 2.0 produced by Thermo Fisher Scientific. The multiple amplification target capture system is 20 μL, add 4 μL 5×Ion AmpliSeq HiFi Master Mix, 10 μL 2×GBTS-InDel1011 multiplex amplification premix primers, 4 μL prepared genomic DNA working solution and 1 μL nuclease-free water, put SimpliAmp TM Thermal c...
Embodiment 3
[0077] Example 3 Construction of Maize DNA Fingerprint Using Multiple Amplification Targeted Capture Sequencing Genotyping InDel Marker Combination
[0078] 1. DNA extraction and quality identification
[0079] 300 maize varieties were selected, and the genomic DNA was extracted by the conventional CTAB method, and the RNA was removed. The quality of the extracted DNA was detected by agarose electrophoresis, and the results showed that the extracted genomic DNA had reached the relevant quality requirements, that is, the agarose electrophoresis showed that the DNA band was single and there was no obvious dispersion; A fluorometer was used to quantify genomic DNA, and the genomic DNA of all corn samples was diluted to 2.5 ng / μL as a DNA working solution for later use.
[0080] 2. Corn DNA fingerprint library construction InDel marker selection
[0081] From the 1011 InDel markers, 384 InDel markers were selected based on the uniform distribution on the chromosome, high stabil...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com