Kit for detecting 33 respiratory pathogens through MALDI-TOF mass spectrum platform
A respiratory and kit technology, which is applied in the field of kits for the detection of 33 respiratory pathogens on the MALDI-TOF mass spectrometry platform, can solve the problems that respiratory pathogens are not easy to quickly realize and cannot meet clinical needs, achieving both flexibility and scalability , low labor cost and detection cost, and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0250] Example 1 A nucleic acid detection kit for detecting 33 kinds of respiratory pathogens
[0251] 1. Composition
[0252] Multiplex PCR amplification primers and single base extension primers, negative quality control (RNasP(+), 33 pathogens (-)) and positive quality control (RNasP(+), FluA(+), SP(+), other pathogens (-)), one-step multiplex PCR amplification reagents (including: multiplex PCR buffer, multiplex PCR enzyme mixture), de-dNTPs reaction reagents (including: phosphatase buffer, phosphodigestive enzyme) and single base extension reaction reagents (including: extension termination mixture, reaction catalytic enzyme, extension buffer), chip reagents (including: desalting resin, mass spectrometry chip).
[0253] Wherein, multiple PCR amplification primers include:
[0254] (1) Primers for specifically amplifying a certain sequence of the novel coronavirus ORF1ab gene,
[0255] F1: 5'-ACGTTGGGATGCCCTGTGGACTTAAGTTTTAC-3' (SEQ ID NO: 1),
[0256] R1: 5'-ACGTTGGAT...
Embodiment 2
[0465] The detection of embodiment 2 samples
[0466] 1. Experimental method
[0467] According to the method of Example 1, the kit of Example 1 was used to detect 3, 10 samples from different sources and known whether they were infected with the above-mentioned 33 kinds of respiratory pathogens, and the sample numbers were 1 to 10.
[0468] 2. Experimental results
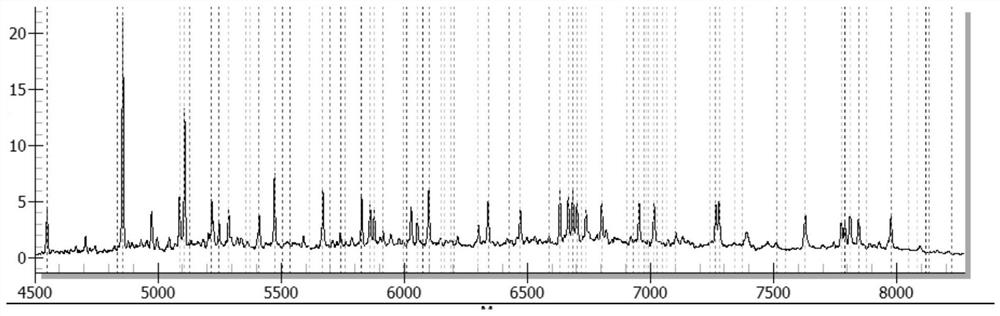
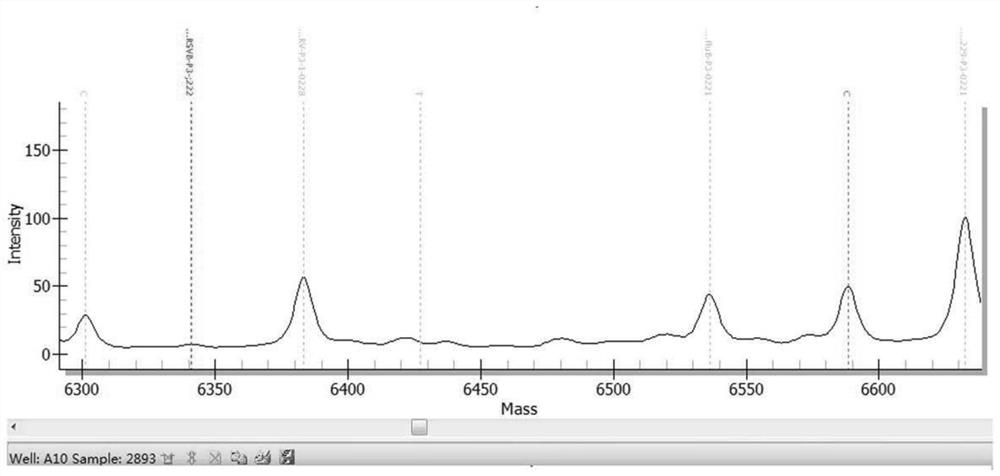
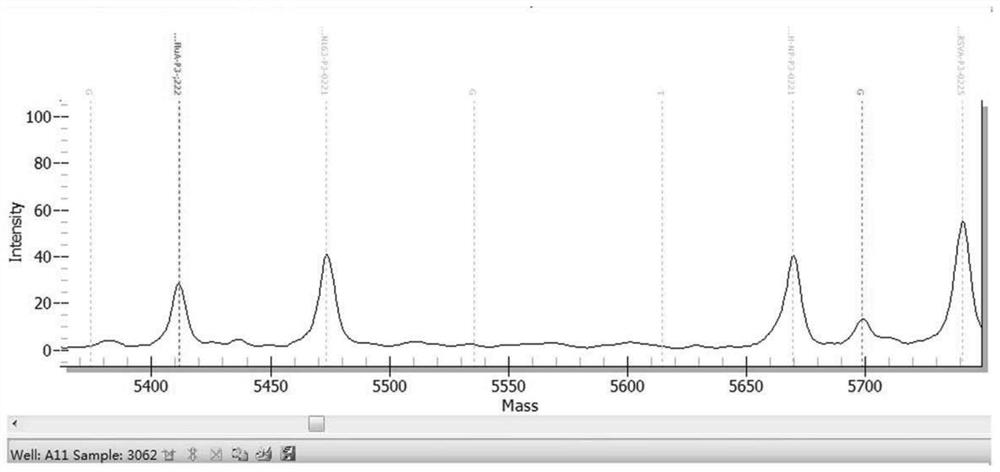
[0469] The results are shown in Table 13. The kit in Example 1 detected 10 samples of known pathogens and the results were consistent with known conditions, with a coincidence rate of 100%. figure 1 Be negative sample representative mass spectrogram in embodiment 2; Figure 2 to Figure 6 It is the positive representative mass spectrum of samples of respiratory syncytial virus B, influenza A virus, mycoplasma pneumoniae, parainfluenza virus I and human coronavirus HCoV-229E in Example 2.
[0470] Table 13 Kit of the present invention detects 10 routine known pathogen sample results
[0471]
[0472]
Embodiment 3
[0473] Sensitivity and accuracy small sample evaluation of embodiment 3 kit
[0474] 1. Experimental method
[0475] 1. Sample selection
[0476] Source and type of samples: 30 cases of nucleic acid samples of clinically confirmed pathogens were selected, including pathogens including novel coronavirus, respiratory syncytial virus (RSVA), and Streptococcus pneumoniae (SP), and 6 cases of plasmid samples, including pathogen detection sites Points are respectively SARS-CoV-2-1ab, LP, SP, HPIV1, HCoV-OC43 and SARS-CoV-2-E, plasmid samples were diluted with 5 gradients (10 2 copies / mL, 10 3 copies / mL, 10 4 copies / mL, 10 5 copies / mL, 10 6 copies / mL) for later use.
[0477] 2. Sample detection
[0478] The above samples were detected using the kit of Example 1 according to the method of Example 1.
[0479] 2. Experimental results
[0480] Table 14:
[0481]
[0482]
[0483] Table 15:
[0484]
[0485] The results are shown in Table 14 and Table 15. The results s...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com