Animal waste metagenome-derived alanine racemase as well as preparation and application thereof
A technology of alanine racemase and metagenomics, which is applied in the field of alanine racemase and its preparation and application, can solve the problems of non-culturable microorganisms, limit the extensiveness, effectiveness and safety of screening, and achieve expansion The effect of using space
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
experiment example 1
[0037] Experimental example 1 Acquisition of alanine racemase gene NC Alr1
[0038] (1) Screening of alanine racemase genes in gibbon fecal microbial metagenomics
[0039] According to the results of gene prediction, functional annotation and secreted protein prediction analysis from the constructed gibbon fecal microbial library, the genes annotated as alanine racemase were screened to obtain the alanine racemase gene NC Alr1. The gene sequence is as follows: Shown in SEQ ID NO.1.
[0040] (2) Cloning of alanine racemase gene NC Alr1
[0041] Take NC Alr 1-F 1 : 5'-TAAGAAGGAGATATACATATGGAATTGATGGATTCAACATTAAAGCGG-3' (SEQ ID NO.3) and NC Alr 1-R 1: 5'-GTGGTGGTGGTG GTGCTCGAGCCCCCTTAAAAGAAGCTC-3' (SEQ ID NO.4) is a primer pair, and PCR amplification is performed using the fecal microbial metagenomic DNA of the western black crested gibbon as a template.
[0042] PCR reaction system (20.0 μL): metagenomic library DNA 0.5 μL, PrimeSTAR Max 10 μL, NCAlr1-F 1 0.5 μL, NCAlr 1-R ...
experiment example 2
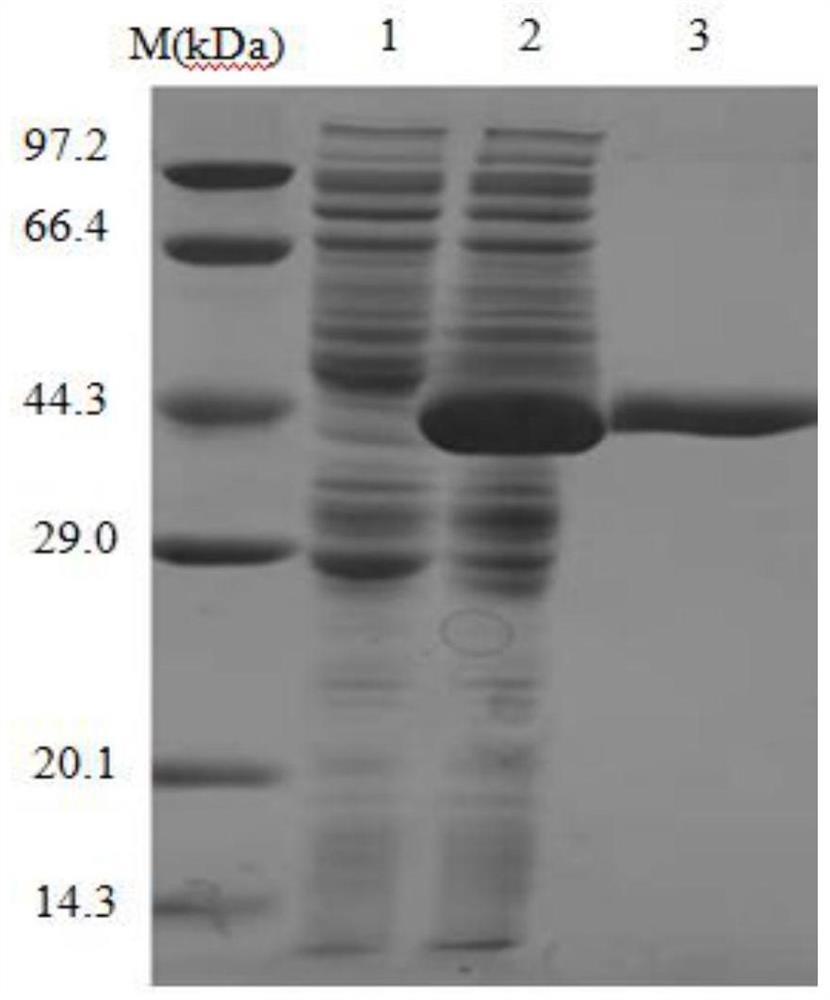
[0045] Experimental example 2 Preparation of alanine racemase NC ALR1
[0046] The alanine racemase gene NCAlr1 prepared in Example 1 was connected to the plasmid pEASY-E2 to obtain the recombinant expression vector pEASY-E2-NCAlr1, and then transformed into E. coli BL21 (DE3) to obtain the recombinant E. coli strain BL21 (DE3) / NCAlr1. Take the Escherichia coli strain BL21(DE3) / NC Alr1 containing the recombinant expression vector pEasy-E2-NCAlr1, and inoculate it in LB (containing 100 μg / mL Amp) culture medium with a 0.1% inoculum size, and cultivate overnight at 37° C. at 180 rpm. Then inoculate the activated bacterial solution into fresh LB (containing 100 μg / mL Amp) culture solution with 1% inoculum amount, and cultivate it at 37° C. at 180 rpm for about 5 to 6 hours (OD 600 After reaching 0.8-1.0), add IPTG with a final concentration of 0.7mmol / L to induce, and culture at 16°C and 180rpm for about 16h. Centrifuge at 5000rpm for 10min to collect the bacteria. After suspen...
experiment example 3
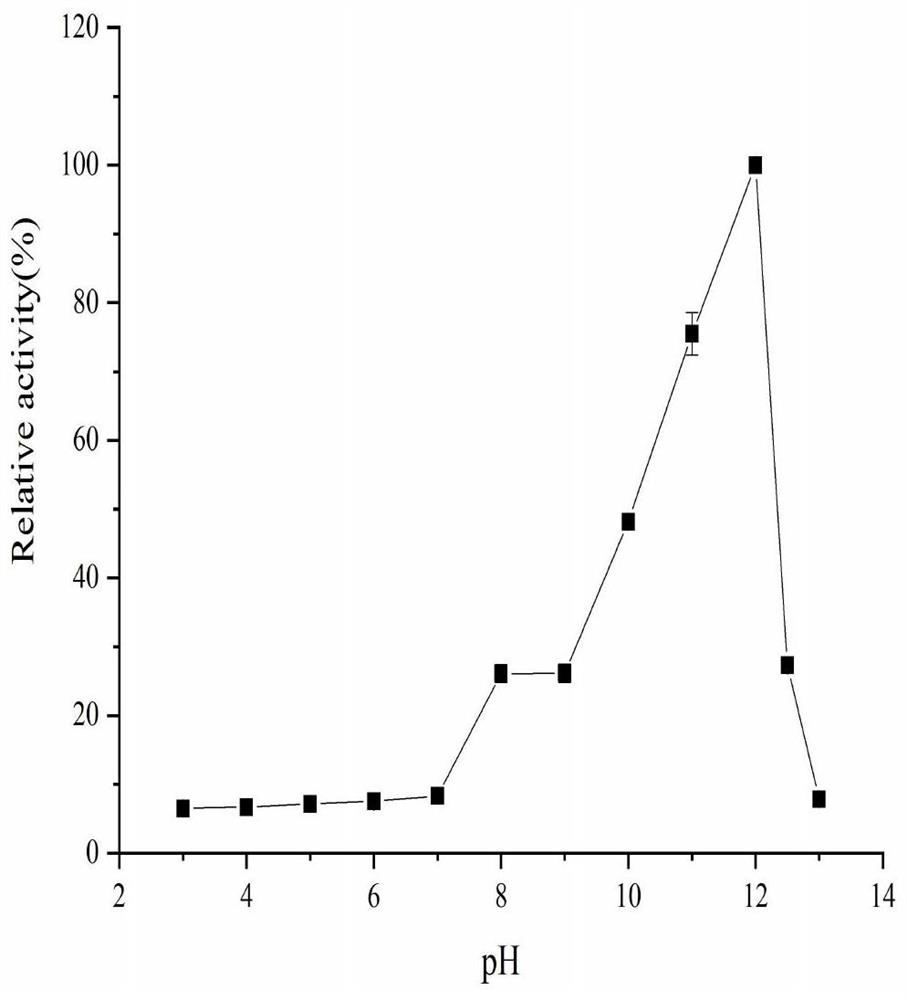
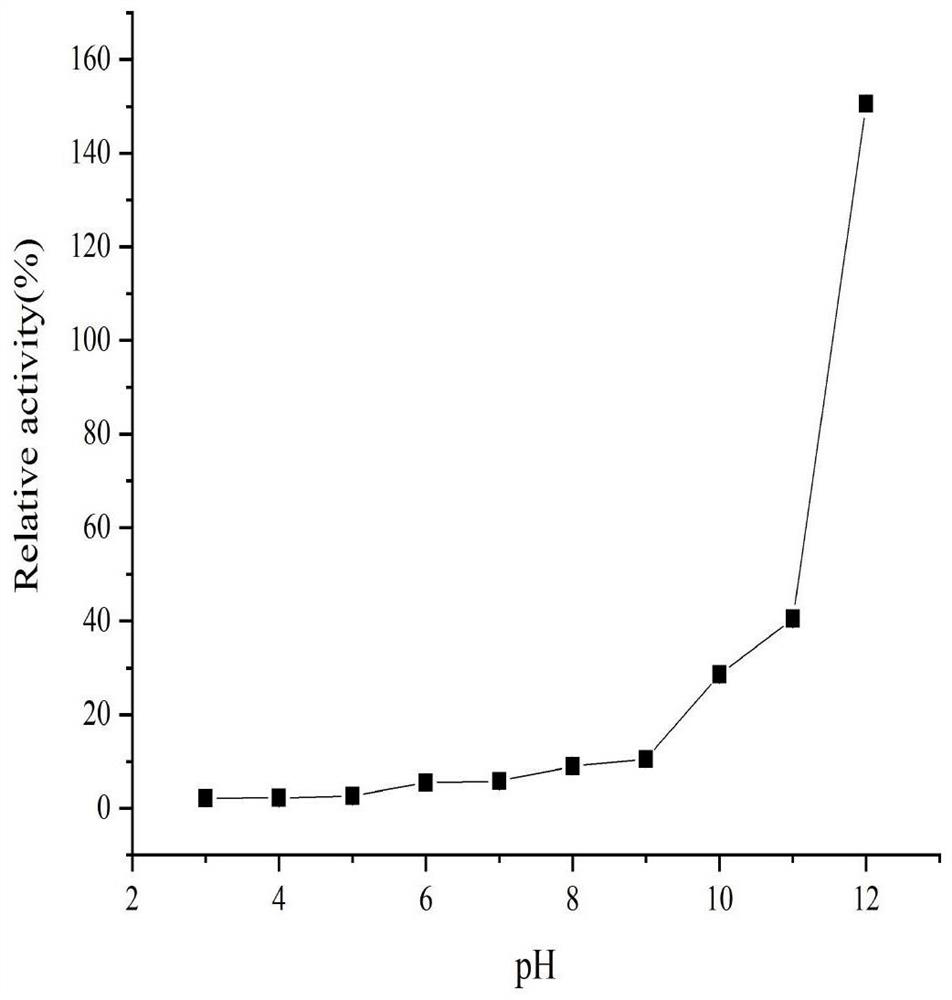
[0048] Experimental example 3 Determination of properties of alanine racemase NC ALR 1
[0049] Enzyme activity determination method refers to Soda K (Microdetermination of D-amino acids and D-amino acid oxidase activity with 3,methyl-2-benzothiazolone hydrazonehydrochloride, 1968) and Lida F et al. (Electrochemical Study of Iodide in the Presence of Phenol and o- Cresol: Application to the Catalytic Determination of Phenol and o-Cresol, 2004): The activity of ALR was determined by a two-step method of racemization reaction and oxidation reaction.
[0050] Racemization reaction: 200 μL reaction system contains 20 mmol / L Beritan-Robinson buffer, 50 mmol / L L-alanine, 0-100 μmol / LPLP (pyridoxal phosphate). Preheat at the optimum temperature for 5 minutes, add an appropriate amount of crude enzyme solution or purified enzyme protein (use boiled inactivated enzyme solution as blank control) to react for 10 minutes, immediately add 25 μL 2mol / L HCl to stop the reaction, and place on...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com