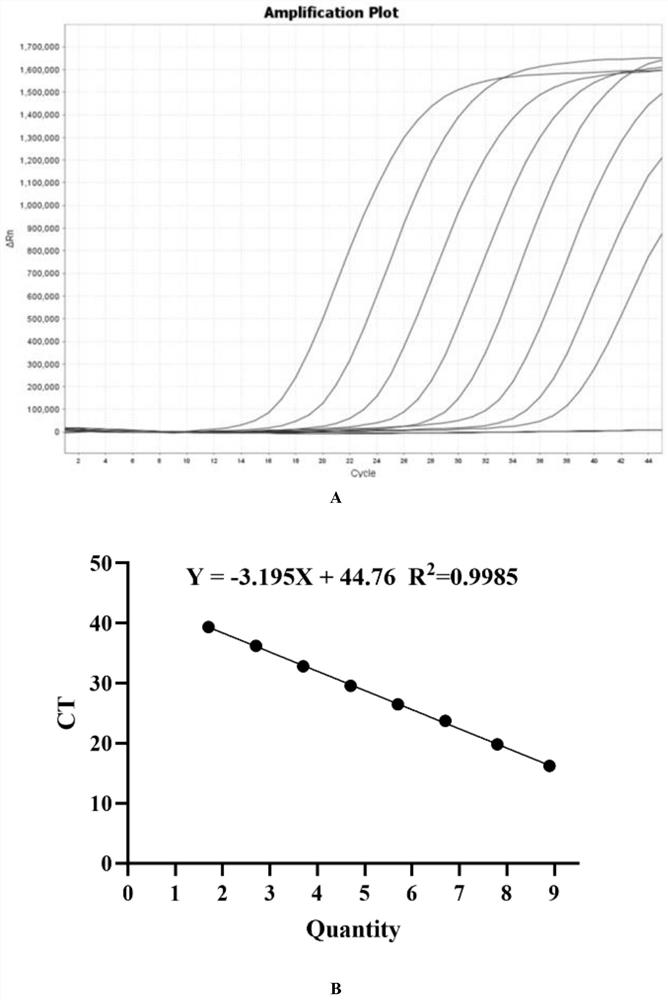
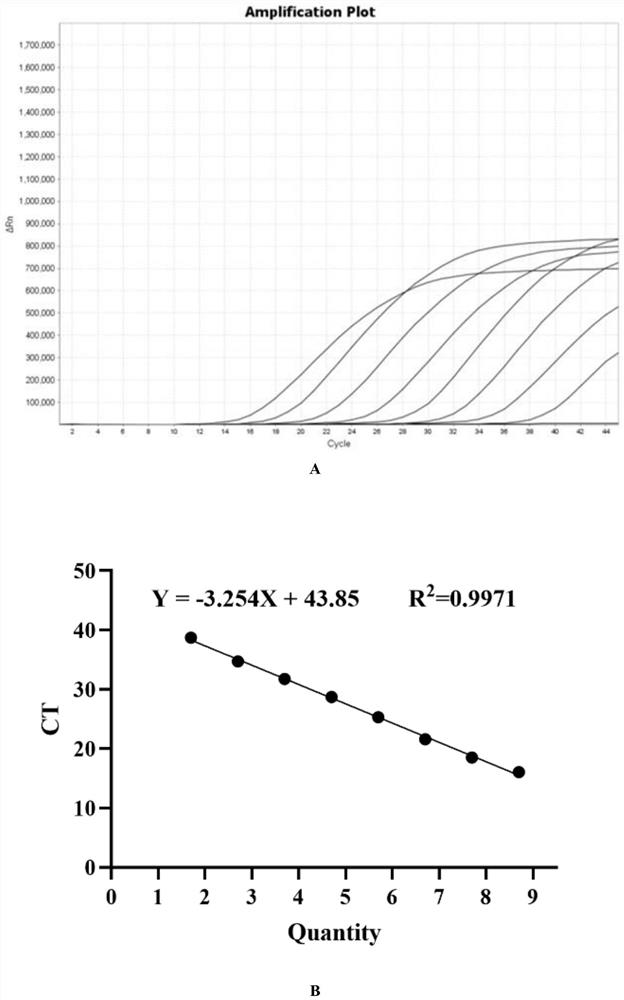
PCR target sequence, primers and probe for detecting infectious SARS-CoV-2 and application of PCR target sequence, primers and probe
A sars-cov-2, target sequence technology, applied in the field of SARS-CoV-2 nucleic acid detection, can solve the difference in PMA binding efficiency, reducing PCR amplification efficiency and sensitivity, GC content, base bias conformation characteristics, etc. problems, to achieve good application prospects, shorten the test cycle, reduce the requirements of scientific research conditions and equipment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Screening and optimization of primers and probes for detection of infectious SARS-CoV-2:
[0027] 1.1 Design of infectious SARS-CoV-2 primers and probes
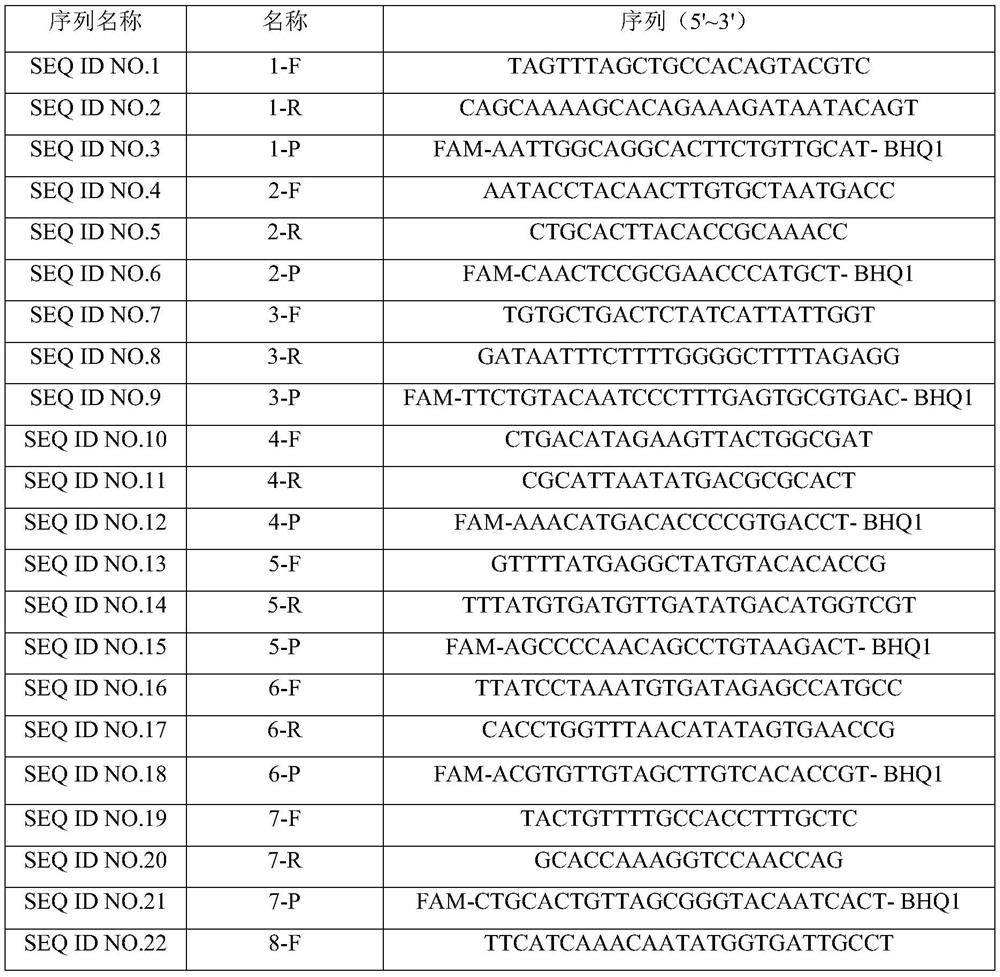
[0028] According to the SARS-CoV-2 (ZY38-1) gene group, 1~15 (SEQ ID NO.1~SEQ ID NO.45) primer probe sets were synthesized, referring to and synthesizing the SARS-CoV-2 detection primer probe released by China CDC Group; China CDC-ORF1ab and China CDC-N (SEQ ID NO.46~SEQ ID NO.51), refer to and synthesize US CDC-N1~N3 (SEQ ID NO. ID NO.52~SEQ ID NO.60). The sequences of the primers and fluorescent probes are as follows (the above primers and probes were synthesized by Shanghai Sangon Biotechnology Co., Ltd.):
[0029]Table 1. SARS-CoV-2 primers and probe sequences
[0030]
[0031]
[0032]
[0033] 1.2qRT-PCR reaction system and reaction conditions:
[0034] Reaction system: Enzyme Mix 2μl, MP buffer 12μl, upstream and downstream primers at a final concentration of 400-1000pM each, probes at a final conc...
Embodiment 2
[0055] Optimization of PMA-Triton X-100-qRT-PCR method for detection of infectious SARS-CoV-2
[0056] 2.1 PMA usage concentration optimization
[0057] Pipette the test samples of inactivated samples in equal volumes, add different concentrations of PMA (final concentrations are 0, 5, 50, 100, 150, 200, 250 μM) and incubate at room temperature for 20 minutes in the dark by vortexing. After the incubation, use PMA- lite TM The LED photolysis instrument photolyzes the sample for 15 minutes. Nucleic acid was extracted with Tiangen automatic nucleic acid extractor and used as a template for qRT-PCR. Considering the use effect and use cost comprehensively, the lowest PMA concentration with large difference in ΔCt or Ct>40 of the sample to be sequenced and no typical amplification curve was selected as the optimal PMA working concentration. The results are shown in Table 5, and the optimal working concentration of PMA is 100 μM.
[0058] Table 5 Optimum PMA Concentration Explor...
Embodiment 3
[0071] Application of multiplex qRT-PCR kit for detection of SARS-CoV-2 infectivity
[0072] This example refers to the "Specific Site Disinfection Technical Plan" (National Health Office Disease Control Letter [2020] No. 156) and "National Food Safety Standard Food Cold Chain Logistics Hygienic Specifications" (GB31605-2020) to simulate high-temperature, alcohol-based disinfectants ( 75% ethanol), chlorine-containing disinfectant (84 disinfectant), and quaternary ammonium salt disinfectant (Germet) treated food packaging surface samples (negative by plaque test, as inactivation completed group), in addition SARS-CoV-2 plaque test-positive virus fluid was used as the non-inactivated group. The optimal detection of infectious SARS-CoV-2 primers and probes and processing methods obtained in Examples 1-2 were applied to the detection of infectious SARS-CoV-2 multiplex qRT-PCR kit.
[0073] Sample collection steps:
[0074] Refer to the sample collection section in "Sampling and...
PUM
| Property | Measurement | Unit |
|---|---|---|
| coefficient of variation | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com