Methods and drugs for targeted editing of RNA based on leaper technology
A targeting and editing technology, applied in DNA/RNA fragments, recombinant DNA technology, other methods of inserting foreign genetic materials, etc., can solve the problems of low editing efficiency, short complementary RNA length, complex chemical modification, etc. Efficient effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
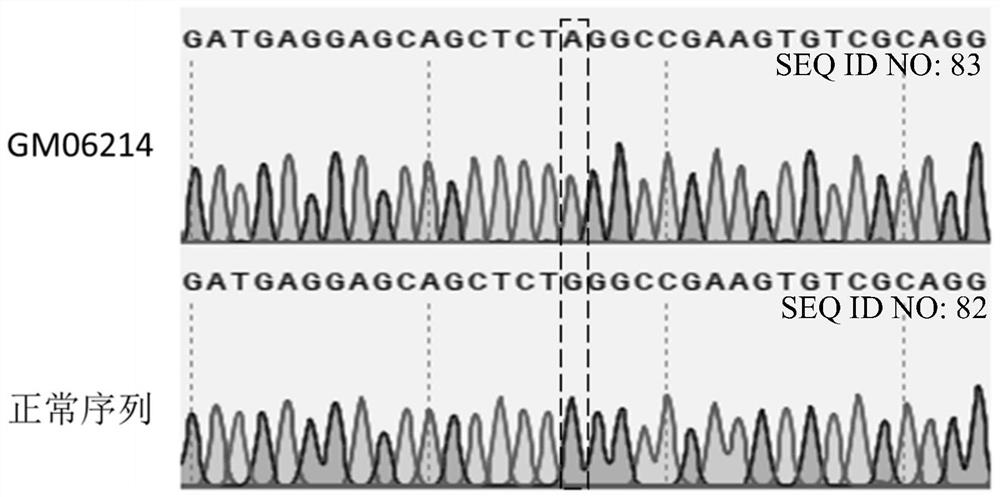
[0260] Example 1: Detection of GM06214 mutant genotype
[0261] Put GM06214 cells into the fibroblast culture medium (ScienCell, FM medium, product number: 2301) containing 15% serum, add 1% fibroblast growth supplement (ScienCell, GFS, product number: 2301), at 37 ℃ , 5%CO 2Cultivate in the incubator for 2-3 days. When the cell confluency reached 90%, it was digested with 0.25% trypsin, and then the digestion was terminated with fibroblast culture medium containing 15% serum. use (TIANGEN Biotech (Beijing) Co., Ltd.) Cellular DNA Extraction Kit (Product No.: DP304-03) was used for DNA extraction according to the operating instructions.
[0262] NCBI-Primer blast (URL: https: / / www.ncbi.nlm.nih.gov / tools / primer-blast / ) was used to design primers for the upstream and downstream sequences of the IDUA target site. SEQ ID NO 1: CGCTTCCAGGTCAACAACAC (forward primer hIDUA-F1); SEQ ID NO 2: CTCGCGTAGATCAGCACCG (reverse primer hIDUA-R1). The PCR reaction was carried out, and the ...
Embodiment 2
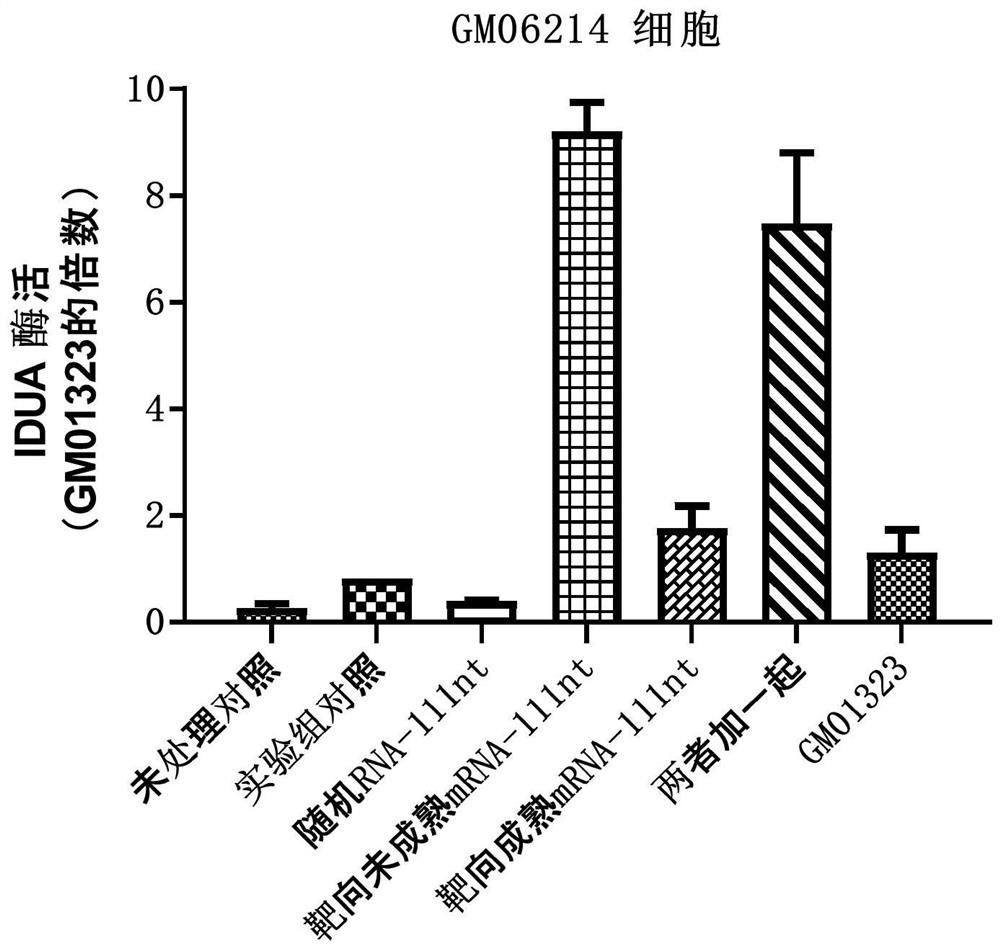
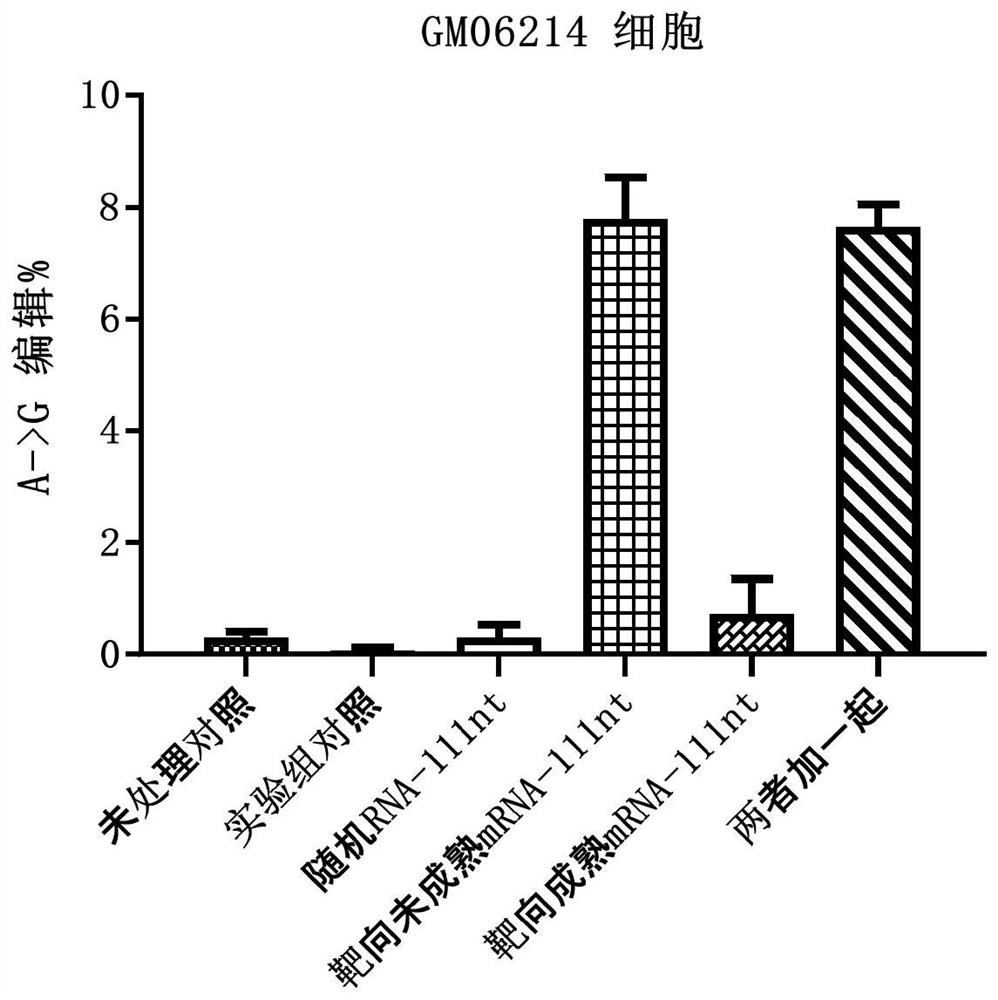
[0263] Example 2: Detection of IDUA enzyme activity and editing efficiency after electrotransfection of arRNA in GM06214 cells
[0264] For the upstream and downstream sequences of the mRNA precursor (pre-mRNA) and mature mRNA target sites after IDUA gene transcription, the following arRNA sequences were designed and synthesized:
[0265] Targeted arRNA against pre-mRNA:
[0266] GACGCCCACCGUGUGGUUGCUGUCCAGGACGGUCCCGGCCUGCGAC ACUUCGGCCCAGAGCUGCUCCUCAUCCAGCAGCGCCAGCAGCCCCAUGGCCGUGAGCACCGGCUU (SEQ ID NO: 3, Pre-55nt-c-55nt);
[0267] Targeted arRNA against mature mRNA:
[0268] GACGCCCACCGUGUGGUUGCUGUCCAGGACGGUCCCGGCCUGCGAC ACUUCGGCCCAGAGCUGCUCCUCAUCUGCGGGGCGGGGGGGGCCGUCGCCGCGUGGGGUCGUUG (SEQ ID NO: 4, m-55nt-c-55nt);
[0269] Randomly designed non-targeting arRNA:
[0270] UACCGCUACAGCCACGCUGAUUUCAGCUAUACCUGCCCGGUAUAAA GGGACGUUCACACCGCGAUGUUCUCUGCUGGGGAAUUGCGCGAUAUUCAGGAUUAAAAGAAGUGC (SEQ ID NO: 5, Random-111nt)
[0271] Wherein, the base in the arRNA for the mutation site ...
Embodiment 3
[0288] Example 3: Detection of IDUA target site editing efficiency after electrotransfection of arRNA on IDUA-reporter cell line
[0289] Such as Figure 3A As shown, a segment containing human (NM_000203.4(IDUA)-c.1205G>A (p.Trp402Ter))(IDUA)_c.1239G>A( p.Trp402Ter) mutation site and the IDUA gene transcript sequence of about 100 bp upstream and downstream respectively to construct a plasmid. The above constructed plasmid was packaged into a virus, infected 293T cells, and after it was integrated into the genome, IDUA-reporter monoclonal cells were screened out. This monoclonal cell only expresses mcherry protein due to the influence of the TAG stop codon where the IDUA target adenosine is located in the inserted sequence, and when the cell is edited by arRNA, it changes from TAG to TGG, so the subsequent GFP protein can be expressed normally . The positive cell ratio of GFP protein can reflect the editing efficiency of arRNA-edited cells. We preferably designed 4 (25nt-c...
PUM
| Property | Measurement | Unit |
|---|---|---|
| recovery rate | aaaaa | aaaaa |
| recovery rate | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com