In-vitro cell platform for pig gene editing sgRNA screening
A technology for gene editing and porcine kidney epithelial cells, applied in the field of in vitro cell platforms, can solve the problem of no efficient gene editing sgRNA in vitro cell platform, etc., and achieve the effects of high efficiency, high throughput, and avoiding ineffective labor.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Example 1 Screening of cells stably expressing Cas9
[0042] Usually in CRISPR-Cas9 gene editing operation, sgRNA and Cas9 gene need to be transferred into cells at the same time. In order to improve the screening efficiency of gene editing sgRNA, the present invention constructs a cell stably expressing Cas9 protein.
[0043] To judge whether the Cas9 gene is successfully transferred into cells and can be stably expressed, it is first necessary to screen the marker gene on the lentiviral vector carrying the Cas9 gene.
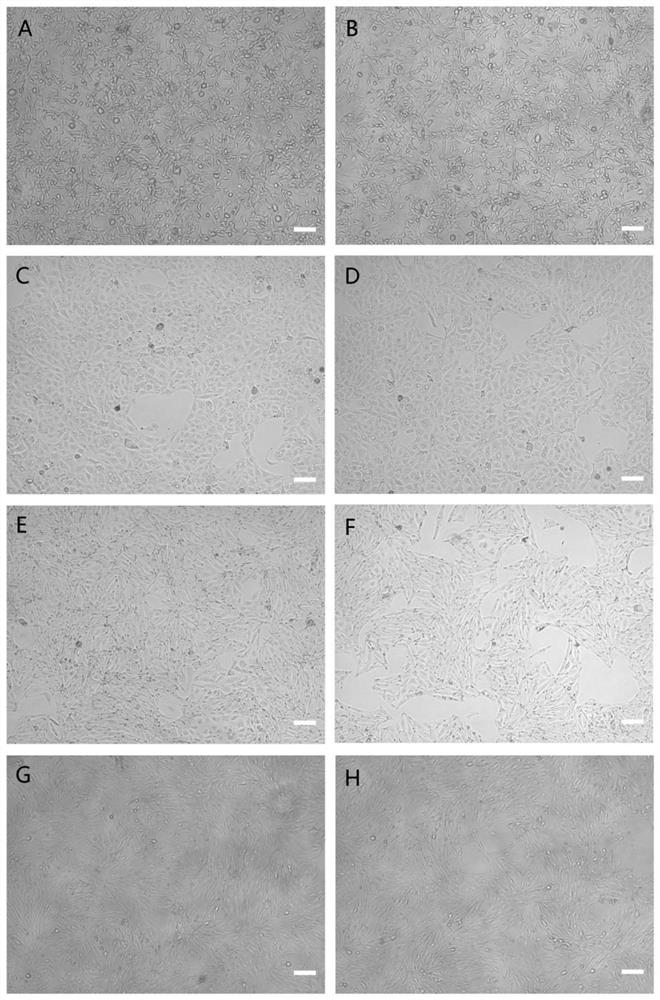
[0044]The pHBLV-gRNA-Cas9-Puro vector used for Cas9 transformation carries a puromycin resistance gene. In order to find pig cells suitable for puromycin screening in this way, the inventors packaged the pHBLV-gRNA-Cas9-Puro vector as a lentivirus and transferred it into 9 target cells [Small Xiang pig skin cells (SSC-S2), Pig skin cells (SSC-S1), minipig kidney cells (SEPfk), minipig lung cells (SEP-L1), porcine kidney epithelial cells (PK15), porcin...
Embodiment 2
[0049] Example 2 RNA-seq (RNA sequencing) analysis of Cas9 stably transfected pig cells
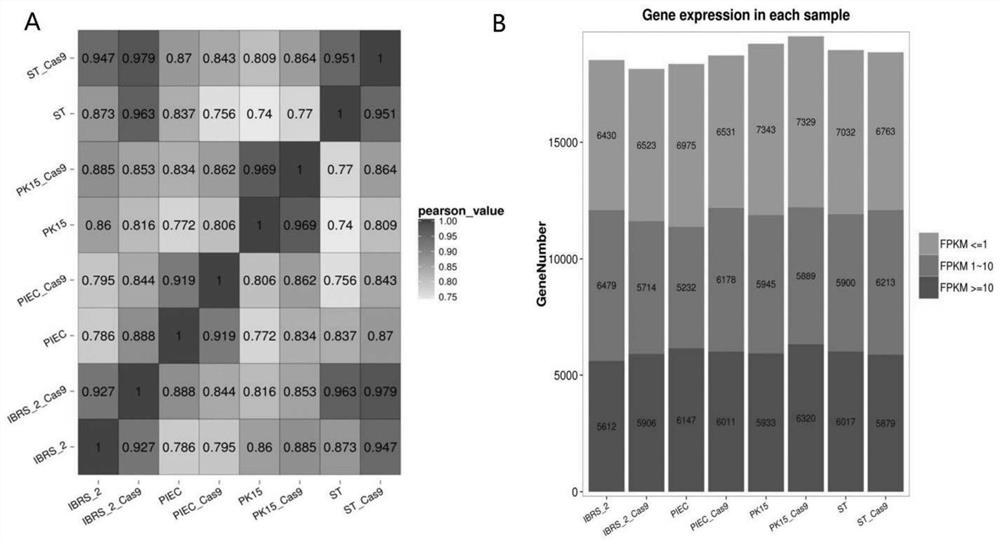
[0050] Based on the Illumina sequencing platform, the inventors performed RNA-Seq on the four cell lines of PIEC / PK15 / IBRS-2 / ST and the corresponding Cas9 stable cells, namely PIEC-Cas9 / PK15-Cas9 / IBRS-2-Cas9 / ST-Cas9 Sequencing and comparative analysis. In order to reflect the correlation of gene expression between samples, this study calculated the Pearson correlation coefficients of all gene expressions between every two samples, and reflected these coefficients in the form of a heat map, as shown in image 3 shown. From image 3 A It can be seen that PIEC and PIEC-Cas9, PK15 and PK15-Cas9, IBRS-2 and IBRS-2-Cas9, ST and ST-Cas9 have the highest correlation, and the Pearson correlation coefficients are all above 0.9, while PIEC / PK15 / The gene expression correlation among the four kinds of IBRS-2 / ST cells was relatively low.
[0051] In order to display the number of genes in differen...
Embodiment 3
[0054] Example 3 Quantitative analysis of proteome of porcine Cas9 stable expression cell line
[0055] Using LC-MS / MS mass spectrometry and TMT labeling quantitative technology combined with bioinformatics analysis, the proteome quantitative analysis of porcine PIEC-Cas9 / PK15-Cas9 / IBRS-2-Cas9 / ST-Cas9 stably transfected cells was carried out, and the stable transfected cells were obtained. Cellular proteome expression. The total number of proteins identified by the four cell lines is 3824, and the protein expression differences of the four Cas9 stably transfected cells can be seen from the heat map of the protein map ( Figure 4 ). In order to facilitate the process-based retrieval, the inventor built a common porcine cell line gene and protein expression retrieval platform PiGenEditer. The link to the web page is: https: / / www.omicsolution.org / wukong / PiGenEditer / , the part of the URL page is as follows Figure 5 .
[0056] Using this platform, you can quickly query the gen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com