Cas12a-CcrRNA system based on regulation and control of nuclease and circular guide RNA and application of Cas12a-CcrRNA system
A nucleic acid and ribozyme technology, applied in the field of analysis and detection, to achieve the effects of high sensitivity, strong anti-interference and good specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Example 1 Based on the Cas12a system regulated by nuclease and circular guide RNA and constructing the technical route of the biosensor
[0050] This application is based on the Cas12a system regulated by nuclease and circular guide RNA and the technical route of constructing a biosensor mainly includes the following steps: (1) C Validate the feasibility of crRNA regulation on the Cas12a system, explore C Effect of crRNA on cis-cleavage and trans-cleavage kinetics of Cas12a protein; (2) Characterization of Cas12a-crRNA binding affinity; (3) Construction of Cas12a system based on nuclease activation; (4) Construction of ATP and two pathogenic bacteria To explore the sensitivity and selectivity of the sensor for target detection, and to use it in the diagnosis of actual clinical samples, to explore the sensitivity and specificity for clinical diagnosis.
Embodiment 2
[0051] Example 2 Exploration C Regulation of the function of Cas12a by crRNA
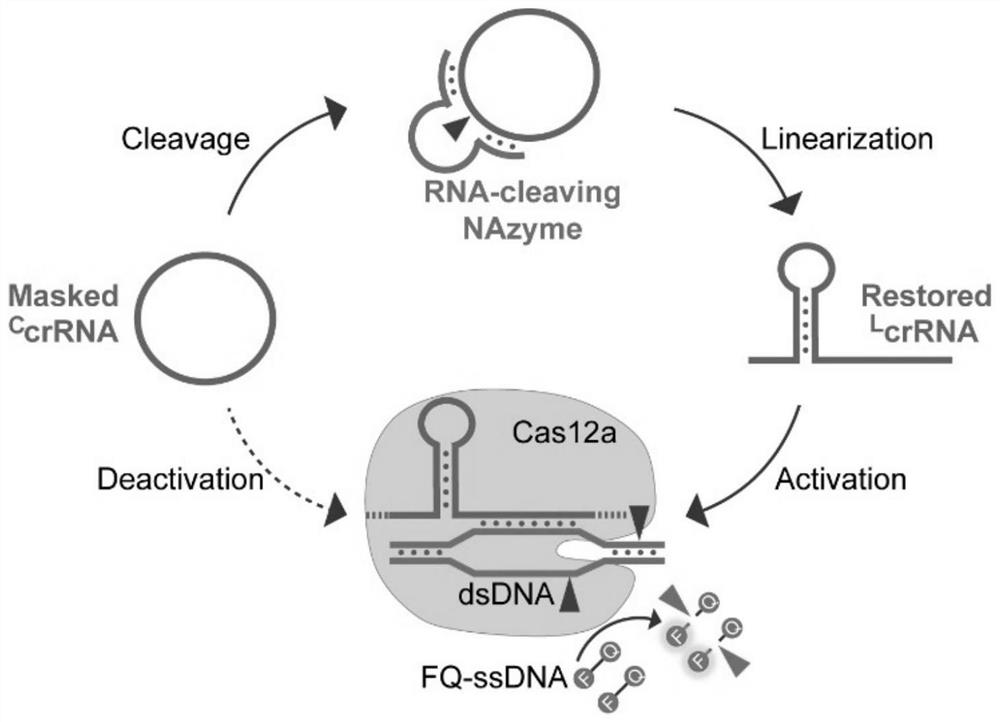
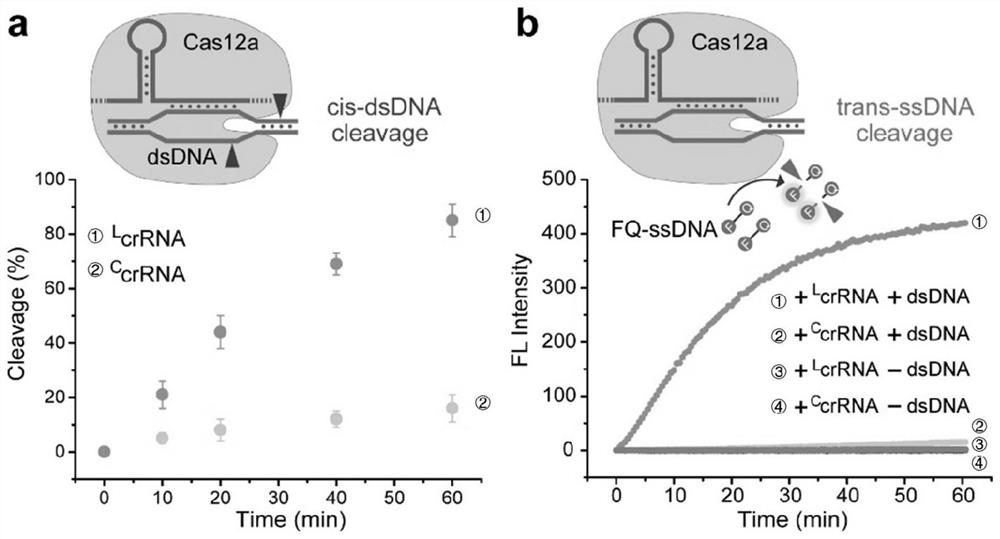
[0052] The present invention builds based on nuclease and C The schematic diagram of the crRNA-regulated Cas12a system is as follows: figure 1 shown, is based on C crRNA can reduce the cleavage activity of Cas12a to DNA, so the present invention explores at first C The regulatory effect of crRNA on the function of Cas12a mainly includes C The effect of crRNA on the reaction kinetics of Cas12a cis-cutting dsDNA and trans-cutting ssDNA.
[0053] (1) C crRNA ii Preparation: 5' phosphorylation L crRNA (such as L crRNA i , L crRNA ii , 100pmol) first with the ligation template (such as LT i , LT ii , 110pmol) and 10μL 10×T4 RNA Ligase2 (T4RL2) buffer (500mM Tris hydrochloric acid, 20mM magnesium chloride, 10mM DTT, 4mM ATP) were mixed, heated to 90°C for 2 minutes, then cooled at room temperature (RT) for 15 minutes; Add 50U of T4RL2, 100U of RNase inhibitor and nuclease-free water to the m...
Embodiment 3
[0058] Example 3 C Affinity characterization of crRNA and Cas12a
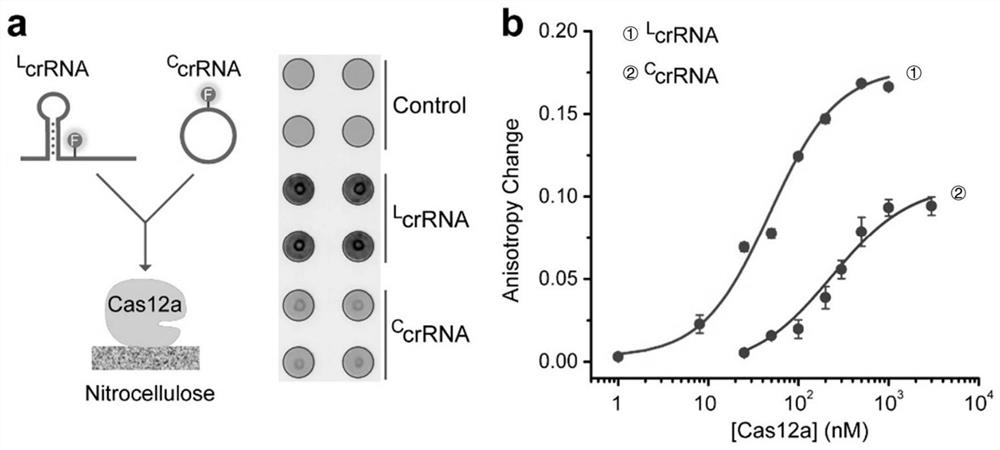
[0059] (1) Dot blot experiment
[0060] A circular test area with a diameter of 4mm was printed on a nitrocellulose membrane (Millipore HF120) by a wax jet printer (Xerox ColourQube 8570N), and heated at 120°C for 2min to melt the wax into the membrane to form a hydrophobic barrier. 1 μL of Cas12a (148ng / uL) was spotted on each test area. After drying at room temperature for 10 minutes, block with 10 μL of 1×BB solution containing 1% BSA for 30 minutes. After washing twice with 20 μL 1×WB, add 10 μL 100 nM FAM-labeled L crRNA ii or C crRNA ii , and incubate for 30 minutes. Place the cardboard in a tank of 2 mL 1×WB to wash for 5 minutes, and after drying, use a fluorescence imager to scan and image.
[0061] Qualitative analysis by dot blot experiments, image 3 a display L crRNA ii The binding affinity to Cas12a is significantly better, while the C crRNA ii have poor binding affinity.
[0062] (2...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com