Purification technique in preparing genetic engineering vaccine of heat shock protein A of recombined Helicobacter pylori
A technology of genetically engineered vaccine and heat shock protein, which is applied in the field of purification process in the preparation of Helicobacter pylori heat shock protein A genetic engineering vaccine, and can solve the problems that the purification process cannot be directly applied
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
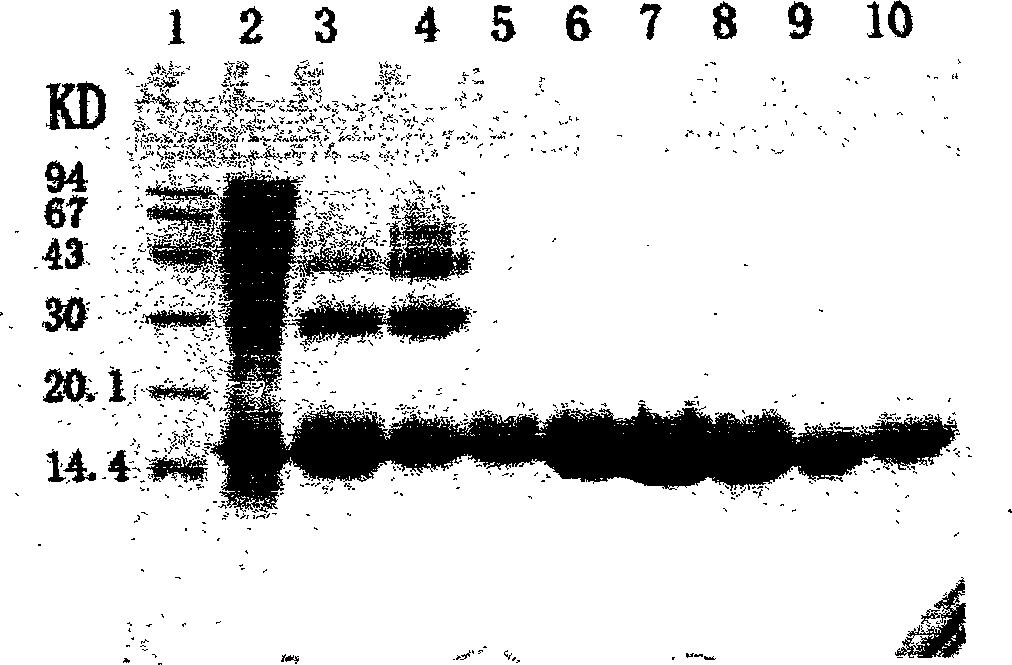
[0031] The recombinant Helicobacter pylori heat shock protein A engineering bacterium constructed by the applicant was fermented at a high density, and the expression rate of the target protein was 23%. The bacteria were collected by centrifugation and purified according to the following steps:
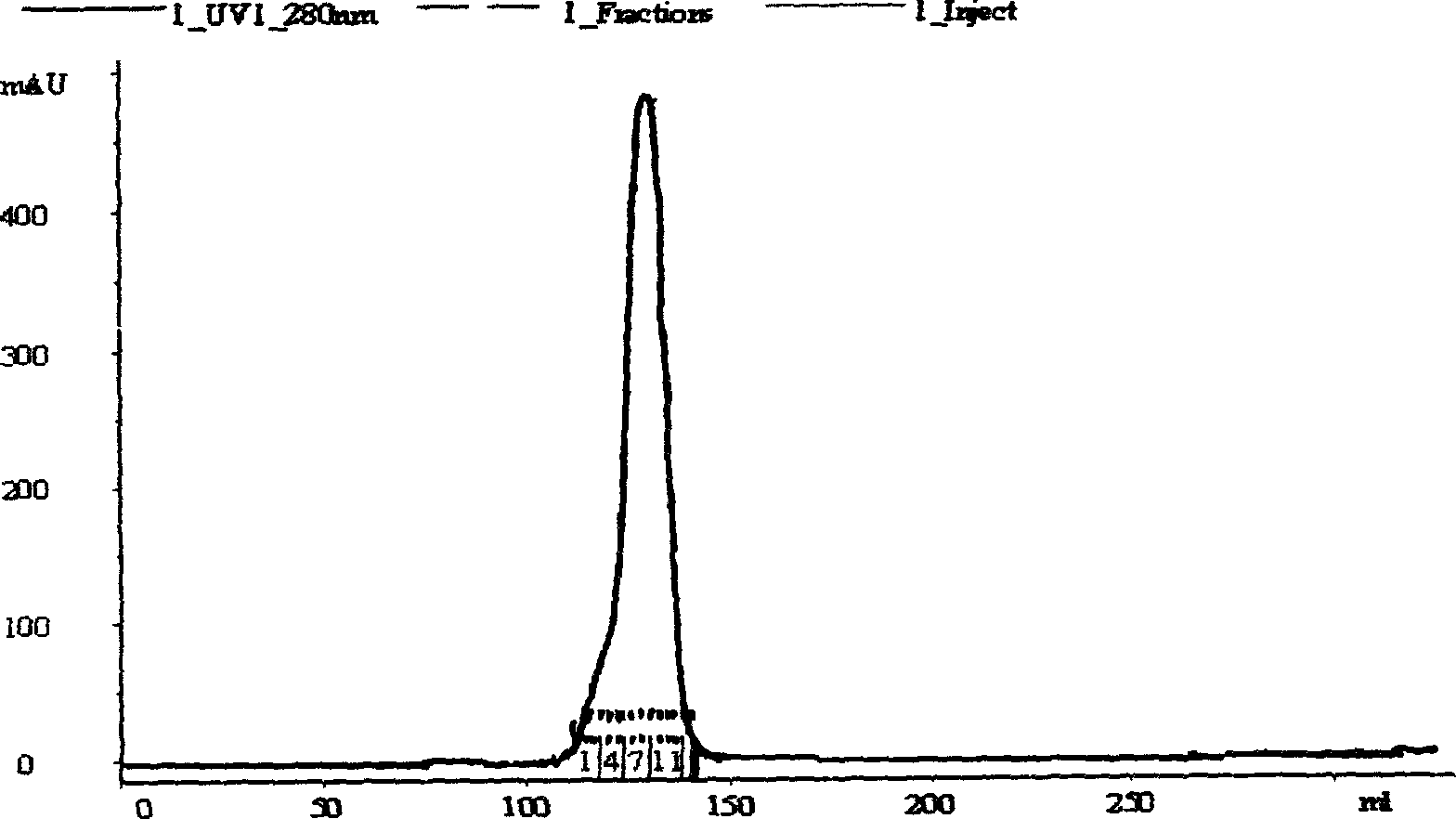
[0032](1) Autoclaving: Suspend 500 g of highly expressed bacteria in TE nickel ion affinity chromatography buffer at a ratio of 1:10 (W / V), pre-cool at 4°C and mix with a cell homogenizer uniform. Use a high-pressure homogenizer to destroy bacteria under the condition of a pressure of 75Mpa (a total of 5 times, until the bacterial cracking rate is greater than 98%). After the bacteria are broken, take a small amount of bacterial liquid smear and stain, and observe the integrity of the cells under a microscope , to ensure that the cells are completely broken, centrifuge at 15,000g for 40min, collect the supernatant and discard the precipitate.
[0033] (2) Filtration: the above supern...
Embodiment 2
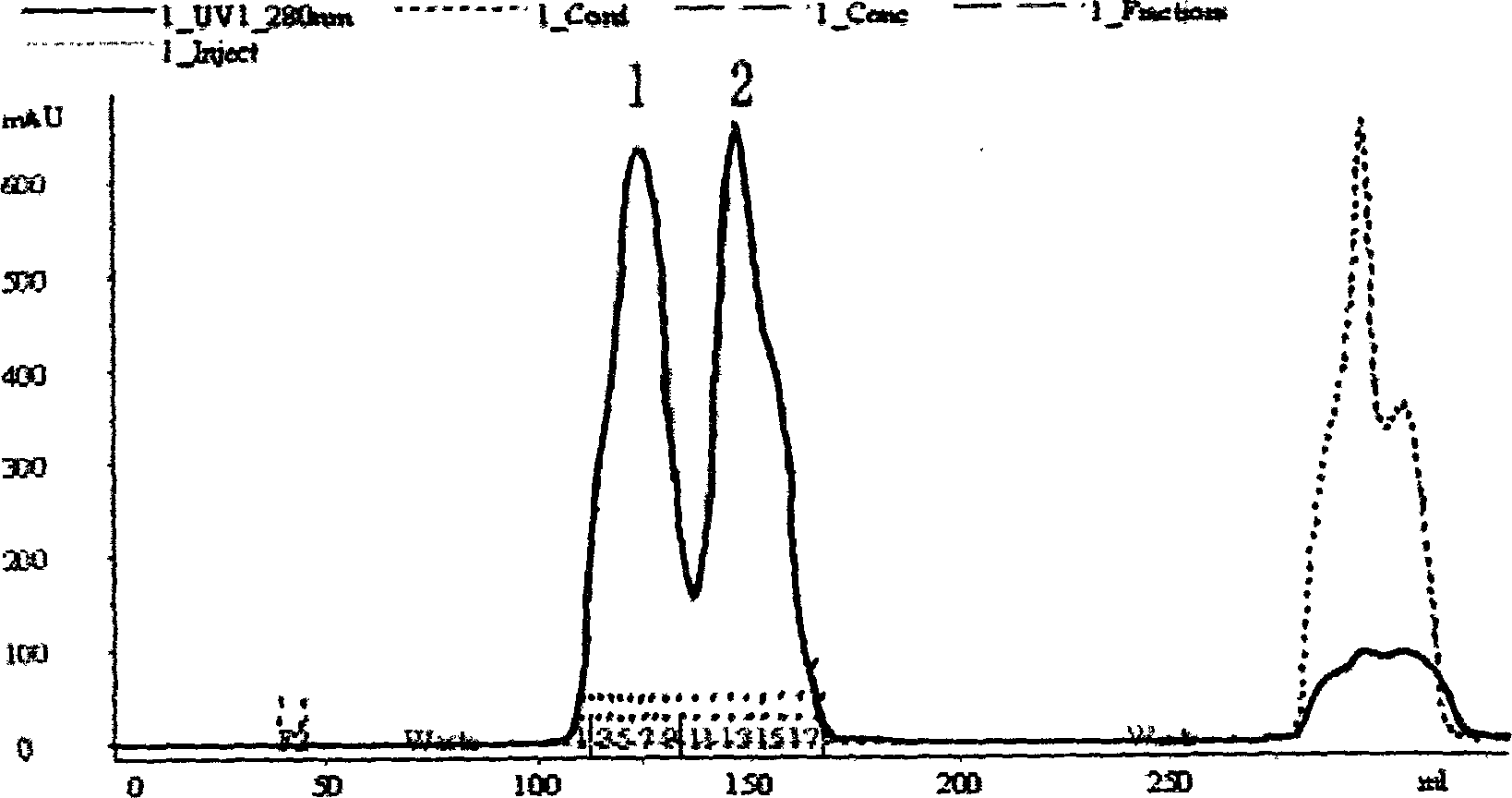
[0039] The sample treatment was the same as in Example 1, and the nickel ion affinity purification column Chelating Sepharose FF was used for preliminary purification, and the purification conditions were the same as in Example 1; 8mol / L urea and 50mmol / L DTT were used to denature and lyse the protein; under denaturing conditions, a gel filtration column Superdex 75 was used Purify (equilibrium buffer: 6mol / L guanidine hydrochloride, 20mmol Tris, pH8.5), and collect the target protein peak. Gel filtration column G-25 was used for denatured protein for desalting and refolding of denaturant, and refolding solution (20mmol NaHCO 3 / Na 2 CO 3 , 20mmol EDTA, pH10), and collect the target protein peak.
Embodiment 3
[0041] Sample treatment and purification conditions are the same as Example 1; denatured cleavage protein adopts 8mol / L urea and 50mmol / LDTT; under denaturing conditions, adopt gel filtration column Superdex 200 to purify (equilibrium buffer: 6mol / L guanidine hydrochloride, 20mmol Tris, pH8. 5), collect the target protein peak; HspA denatured protein, use the nickel ion affinity chromatography column Chelating Sepharose HP to carry out on-column renaturation, the method is that the chromatography column is equilibrated with equilibrium solution (8mol / L urea, 20mmol Tris, pH 8.5) Afterwards, load the denatured target protein sample, use refolding solution (20mmol / L PBS, 2mmol GSH, 0.2mmol / L GSSG, 0.15mol / L NaCl) to slowly gradient elute the column until urea is completely removed, use imidazole buffer (20mmol / L PBS, 0.5mol / L imidazole, pH8.5) to elute the target protein and collect the protein peak.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com