Barbadosnut salt induced transcription factor and its coding gene and uses
A transcription factor, salt-induced technology, applied in the application, genetic engineering, plant genetic improvement and other directions, can solve the problems of plant stress resistance, plant stress resistance has not been comprehensively improved, single functional gene and other problems, to achieve high practical Application value, effect of improving stress resistance and broad application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Embodiment 1, the acquisition of the complete cDNA sequence of the salt-induced transcription factor gene JcERF of Jatropha curcas
[0048] The acquisition of the full-length cDNA sequence of the Jatropha curcas salt-induced transcription factor gene JcERF comprises the following steps:
[0049] 1. Cloning of the 3' end sequence of the salt-induced transcription factor gene JcERF of Jatropha curcas
[0050] 1. Plant material processing and total RNA extraction
[0051] First take Jatropha curcas seedlings as material, extract total RNA after being treated with 300mM NaCl solution for 12 hours, and carry out 1% agarose gel electrophoresis detection to it, and the detection result is as follows: figure 1 As shown, the extracted total RNA has two obvious electrophoresis bands, which are 28s RNA and 18s RNA from top to bottom, indicating that the total RNA with high purity and completeness has been obtained.
[0052] 2. Cloning of the 3' end sequence of the salt-induced t...
Embodiment 2
[0069] Example 2, Bioinformatics analysis of JcERF and its encoded protein
[0070] 1. Sequence analysis of JcERF gene and prediction of structure and function of its encoded protein
[0071] Utilize DNAMAN software to carry out bioinformatics analysis to the full-length cDNA sequence of JcERF that embodiment 1 obtains, and its structural representation is as follows Figure 5 As shown (the single line indicates the 3'-UTR; the box indicates the open reading frame, which contains two functional motifs, the black box indicates the AP2 domain, and the hatched line on the right indicates the acidic activation region), the total length of the sequence is 759bp, from 5 The 1st-759th base at the 'end is ORF, which encodes a protein consisting of 253 amino acid residues. It is estimated that its molecular weight is 27.474kDa, and its isoelectric point pI value is 9.09. Using the SMART tool to predict the function of the deduced amino acid residue sequence, the result is that the pro...
Embodiment 3
[0076] Embodiment 3, the structural analysis of JcERF genomic DNA
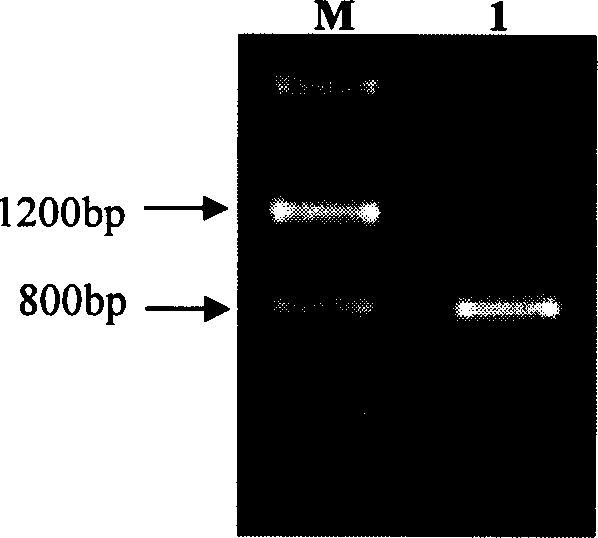
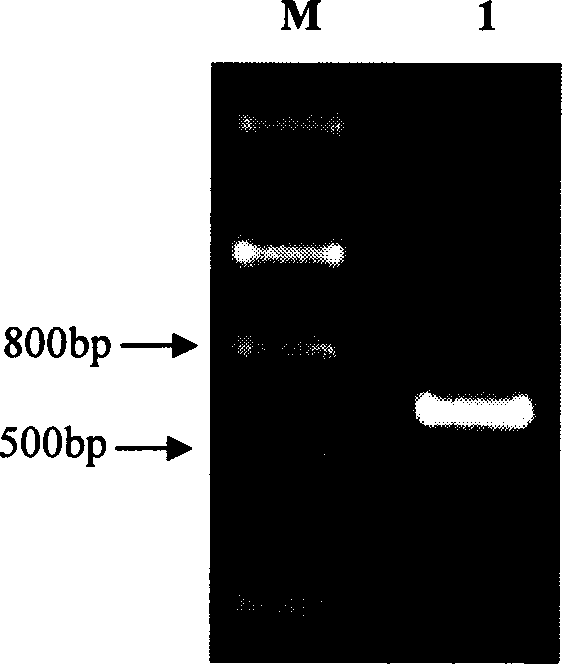
[0077] Extract the genomic DNA of the Jatropha curcas seedling that is processed through 300mM NaCl solution in the embodiment 1 and take this as template, under the guidance of primer JcERFW-1 and and JcERFW-2, carry out the structure of PCR amplification analysis JcERF gene, reaction finishes Afterwards, the PCR products were detected by 1% agarose gel electrophoresis, and the detection results were as follows: Figure 9 As shown (swimming lane M is the molecular weight standard Marker III, and swimming lane 1 is the PCR amplification product), as a result, a specific band with a length of about 800 bp was amplified by PCR. Then use the cDNA of JcERF as a template, carry out PCR amplification with the above-mentioned same primers, after the reaction finishes, carry out 1% agarose gel electrophoresis detection to the PCR product, the detection result is as follows Figure 9 As shown (swimming lane M is the m...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com