RNA affinity media and preparation method thereof
A medium and affinity technology, applied in the field of molecular biology, can solve problems such as the influence of RNA and its binding protein, change the spatial configuration of RNA molecules, etc., and achieve the effect of strong selectivity and easy preparation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
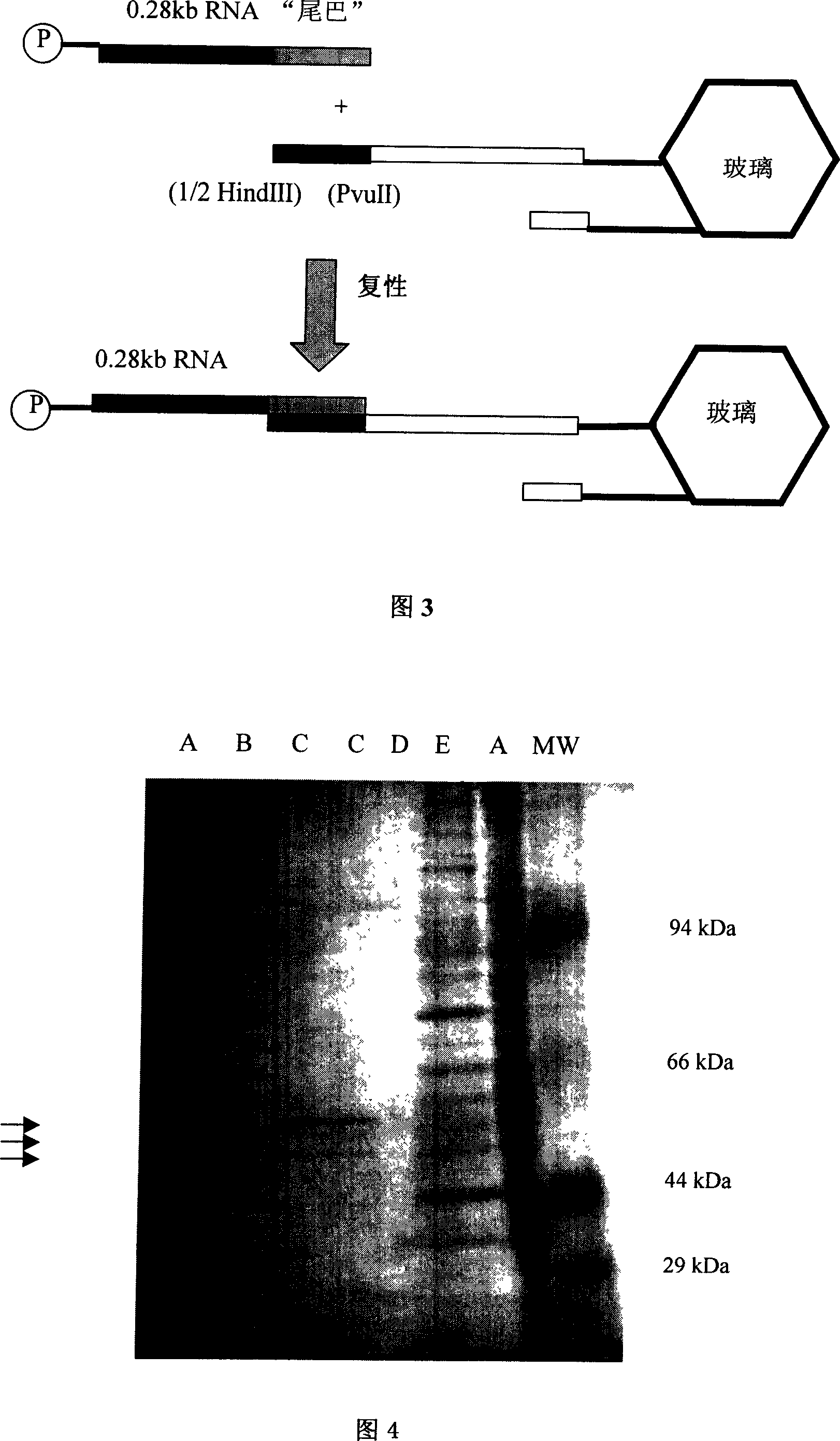
[0035] Example 1, preparation of target RNA with "tail"
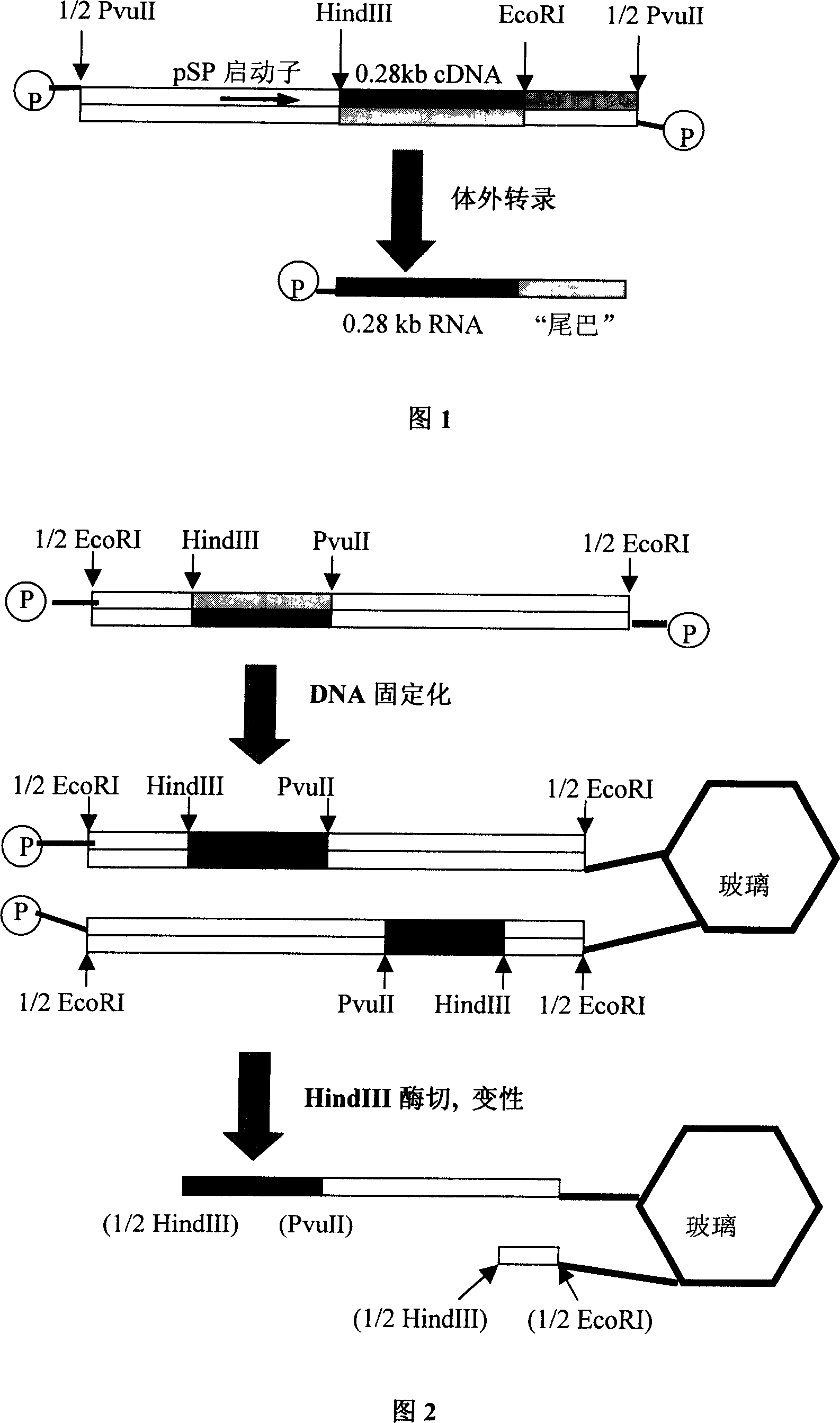
[0036]1.1. The 3'-untranslated region of the mRNA of nuclear transcription factor C / EBPβ is cDNA with a length of 0.28kb. For the specific nucleotide sequence, see Liu Dingqian et al. (Liu Dingqian, Zhu Lihua, Chen Zhenzhen, Noda Liang, Guo Lihe, Li Zaiping, with The nucleotide sequence of a cDNA clone of anti-cancer gene activity, the literature of Biochemistry and Biophysics Acta 1991; 23 (3): 246-250), it is forward cloned into the multi-cloning position of plasmid pSP64 (Promega company) point. For the specific operation of cloning, please refer to Liu Dingqian et al. Recombinant plasmid pSP64 / 0.28. like figure 1 As shown, downstream of the multiple cloning site of the plasmid, the recombinant plasmid pSP64 / 0.28 was digested with PvuII to linearize the recombinant plasmid pSP64 / 0.28 to obtain a linear DNA containing the pSP promoter and 0.28kb cDNA.
[0037] 1.2. Use a kit (RiboMax large scale RNA production kit...
Embodiment 2
[0038] Embodiment 2, the treatment of glass frit carrier and aminosilanization
[0039] Ordinary glass powder (50-100 mesh) is washed with chloroform to remove oil stains, then fully washed with 5N NaOH and 5N HCl in turn, rinsed with a large amount of distilled water to remove residual HCl, and dried for later use.
[0040] Clean glass powder is soaked in 50% acetone solution of 3-aminopropyltriethoxysilane (purchased from Sigma Company), soaked for half an hour at room temperature, and then fully wash away residual 3-aminopropyltriethylsilane with acetone. Oxysilane, dried at 50°C for later use.
Embodiment 3
[0041] Embodiment 3, immobilizing DNA on the surface of aminosilanized glass frit carrier
[0042] 3.1, such as figure 2 As shown, the plasmid pSP65 (Promega) was digested and linearized with EcoRI, and the specific conditions were as follows:
[0043] Plasmid pSP65 20mg 2mL
[0044] 10 times concentration EcoRI buffer 250μL
[0045] 0.1% bovine serum albumin 250μL
[0046] EcoRI 1000 units (100 μL)
[0047] Incubate at 37°C overnight (12-16 hours).
[0048] The next day, take 2 μL of electrophoresis to check whether the digestion is complete; if the digestion is not complete, add 1000 units of EcoR I, continue to incubate for 4 hours, and check by electrophoresis to confirm that the digestion is complete.
[0049] 3.2. Extract the reaction solution with complete digestion with an equal volume of Tris-HCl (pH8) saturated phenol / chloroform (v / v 1:1) once, extract once with chloroform, take out the water phase, and add twice the volume of absolute ethanol , centrifuge at ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com