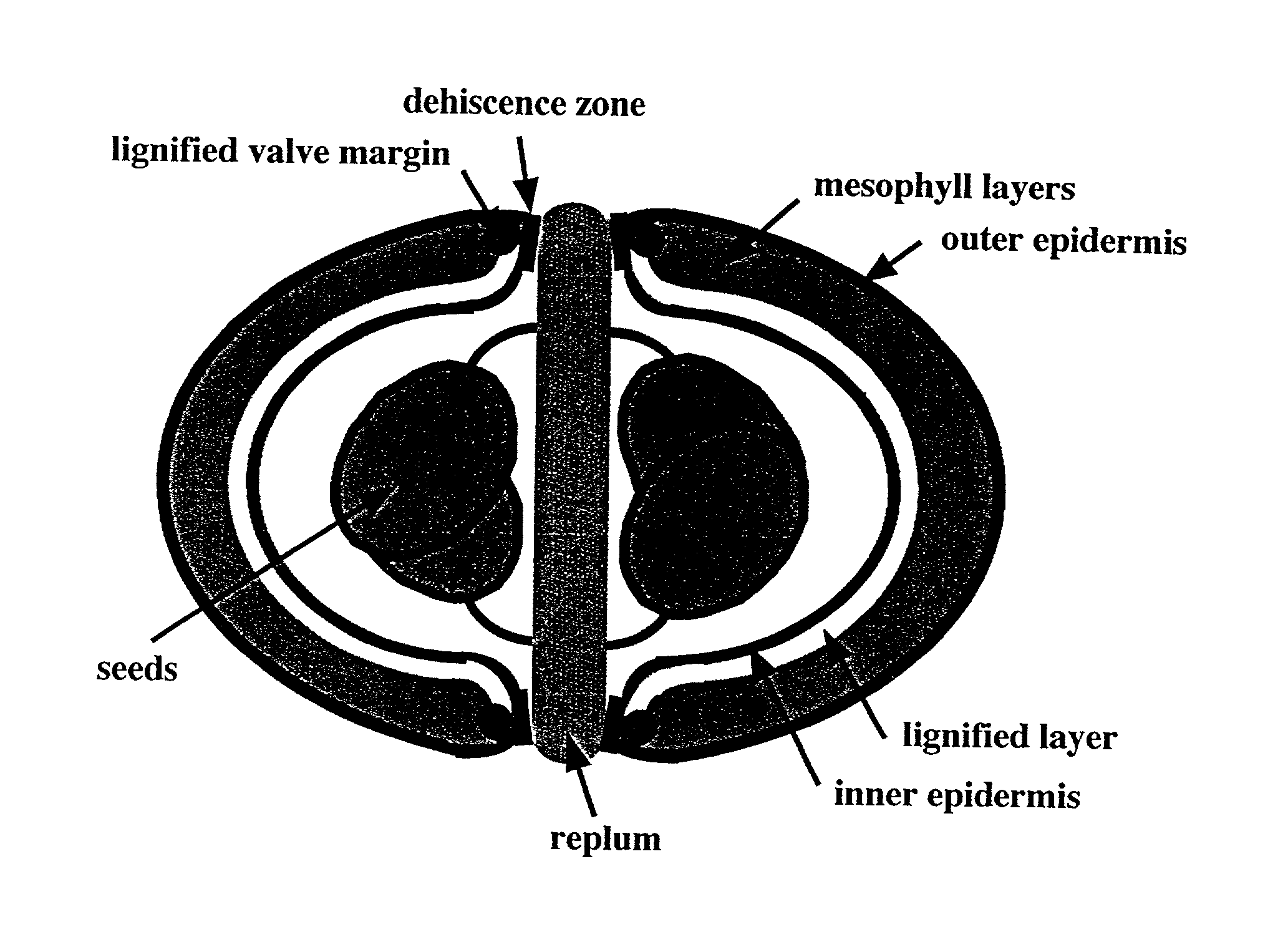
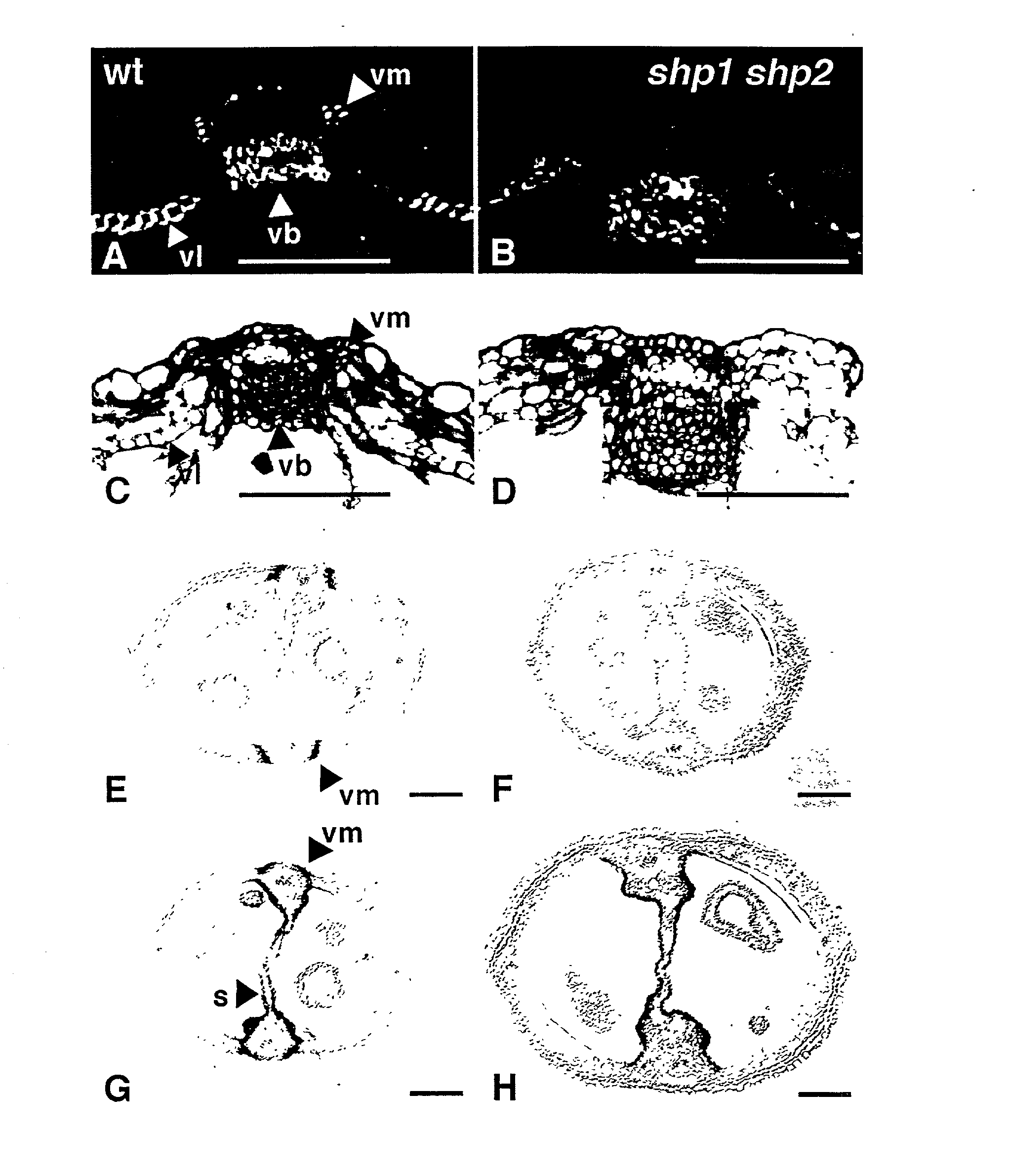
Selective control of lignin biosynthesis in transgenic plants
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example i
Production of a 35S::AGL8 Transgenic Arabidopsis Plant Displaying Reduced Lignification
[0134] This example describes methods for producing a transgenic Arabidopsis plant characterized by reduced lignification due to constitutive AGL8 expression.
[0135] Full-length AGL8 was prepared by polymerase chain reaction amplification using primer AGL8 5-.gamma. (SEQ ID NO:9; 5'-CCGTCGACGATGGGAAGAGGTAGGGTT-3') and primer OAM14 (SEQ ID NO:10; 5'-AATCATTACCAAGATATGAA-3'), and subsequently cloned into the SalI and BamHI sites of expression vector pBIN-JIT, which was modified from pBIN19 to include the tandem CaMV 35S promoter, a polycloning site and the CaMV polyA signal. Arabidopsis was transformed using the in planta method of Agrobacterium-mediated transformation essentially as described in Bechtold et al., C.R. Acad. Sci. Paris 316:1194-1199 (1993), which is incorporated herein by reference. Kanamycin-resistant lines were analyzed for the presence of the 35S-AGL8 construct by PCR using a prime...
example ii
Production of an Arabidopsis agl1 agl5 Double Mutant Displaying Reduced Lignification
[0140] This example describes the production of an agl1 agl5 double mutant displaying reduced lignification.
[0141] A. Production of an agl5 Mutant by Homologous Recombination
[0142] A PCR-based assay of transgenic plants was used to identify targeted insertions into AGL5 as described in Kempin et al., Nature 389:802-803 (1997), which is incorporated herein by reference. The targeting construct consisted of a kanamycin-resistance cassette that was inserted between approximately 3 kb and 2 kb segments representing the 5' and 3' regions of the AGL5 gene, respectively. A successfully targeted insertion produces a 1.6 kb deletion within the AGL5 gene such that the targeted allele encodes only the first 42 of 246 amino acid residues, and only 26 of the 56 amino acids comprising the DNA-binding MADS-domain. The recombination event also results in the insertion of the 2.5 kb kanamycin-resistance cassette wit...
example iii
Production of an agl8 Mutant Arabidopsis Plant Displaying Enhanced Lignification
[0157] This example describes methods for producing a non-naturally occurring plant characterized by enhanced lignification due to suppression of AGL8 expression.
[0158] A. Production of an agl8 Mutant
[0159] A mutation designated ful-1 was identified using large scale insertional mutagenesis with enhancer and gene trap Ds transposable elements (Sundarsen et al., supra, 1995; Springer et al., Science 268:877-880 (1995), each of which is incorporated herein by reference). This system utilized the maize Ac / Ds transposable elements and the reporter gene GUS. Transposition events were selected and screened for reporter gene expression patterns and mutant phenotypes. The ful-1 mutant was identified in the F3 progeny of an enhancer trap line. Backcrossing to wild type Landsberg erecta confirmed that ful-1 is a recessive mutation.
[0160] To address whether the ful-1 mutation was caused by insertion of the Ds-GUS e...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Selectivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com