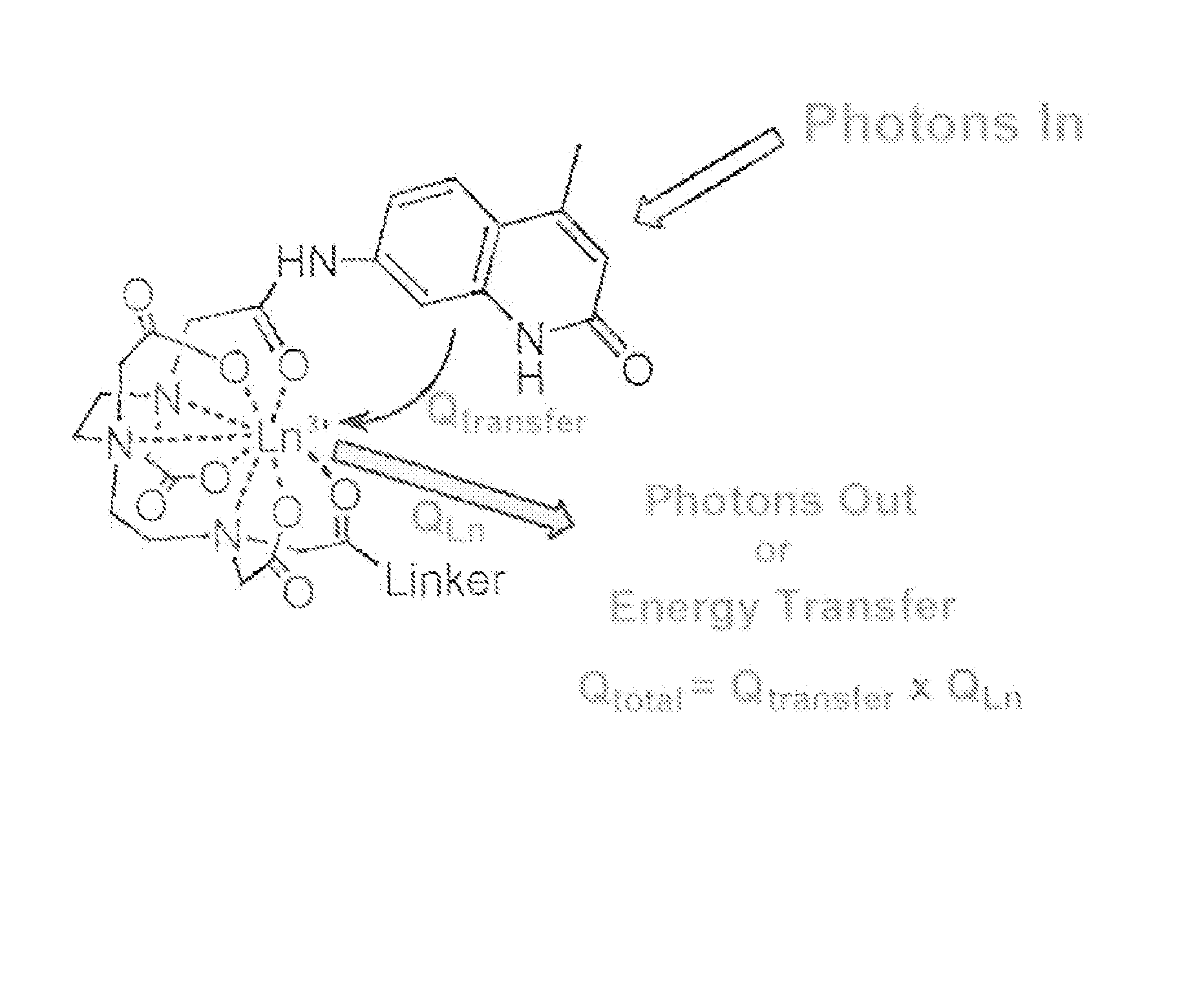
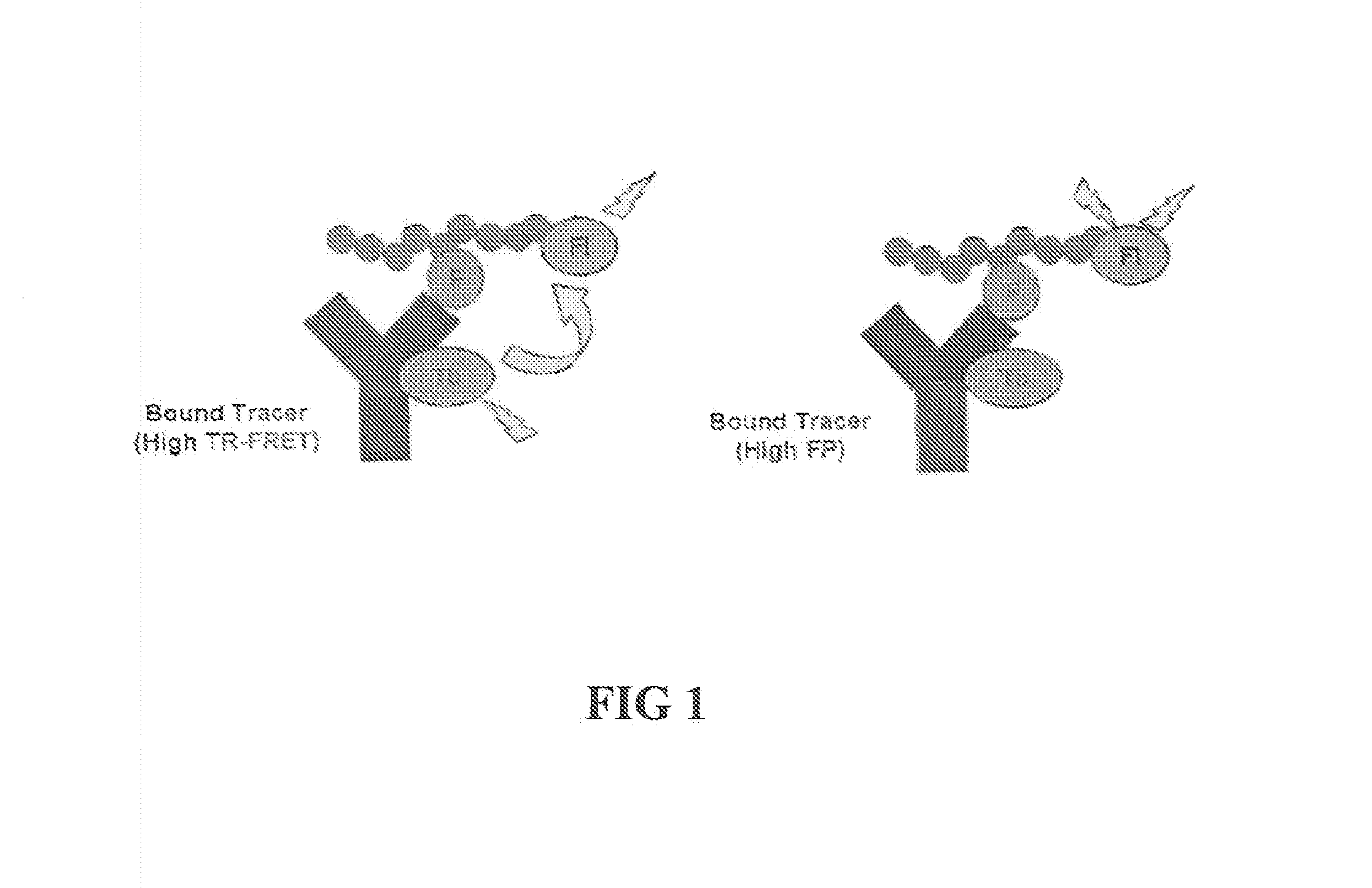
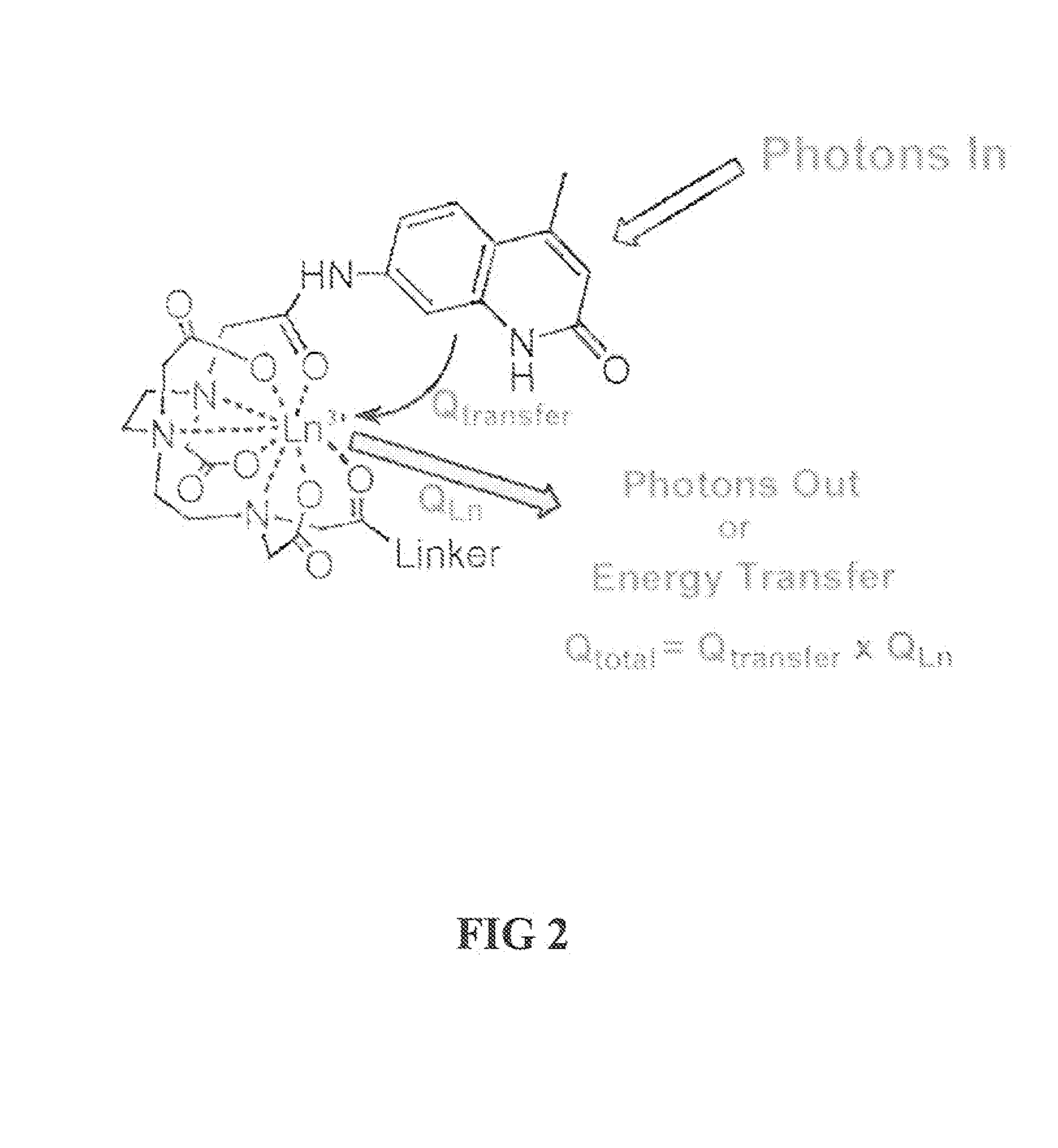
Kinase and ubiquination assays
a kinase and ubiquitination technology, applied in the field of fluorescent molecule and luminescent metal complex assays, can solve the problems of limiting the sensitivity of luminescence-based assays, detection of false positives or false negatives in drug or compound screens, etc., to reduce interference, enhance/up-regulate or modulate an activity, and inhibit the effect of interferen
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Labeling of an Antibody with a Luminescent Metal Chelate
[0294] 1 mg purified PY72 (anti-phosphotyrosine) IgG antibody, an antibody that preferentially binds amino acid sequences containing phosphorylated tyrosines (e.g., sequences phosphorylated by protein tyrosine kinases (PTKs)) and was dialyzed for 1.5 hours in a 100 mM sodium bicarbonate buffer, pH 9.5, using a 12-14,000 MWCO dialysis membrane. (PY72 hybridoma cells were obtained from the Salk Institute; the immunogen was phosphotyrosine conjugated to KLH. Ascites were produced by Harlan Bioproducts for Science, Indianapolis Ind. Ascites were purified with a protein G column (Pierce). Purified antibody is also available from Covance, Berkeley Calif. (Part # MMS414P).) The antibody was then removed from the dialysis membrane and concentrated to 48.8 uM (7.3 mg / mL) using a Centricon YM50 (Millipore) concentrator. 100 uL of this antibody solution was diluted to 5 mg / ml (33.4 uM) into the labeling reaction which consisted of 10 mM ...
example 2
Binding Curve Experiment Between Protein Tyrosine Kinase Product Tracer (PTK Tracer) and Anti-PTK Product (PY72) Antibody
[0296] A direct binding curve (showing luminescent metal chelate-labeled PY72 antibody binding to fluorescent acceptor labeled tracer) was generated by incubating serial dilutions of the labeled antibody (10 nM to 9.8 pM in two fold dilutions) with 1 nM fluorescent acceptor-labeled tracer (PTK labeled tracer; sequence F-ADE(pY)LIPQQS, where F is fluorescein and pY is a phosphorylated tyrosine, SEQ ID NO:1; note that the tracer is a phosphorylated tyrosine derivative of a protein tyrosine kinase (PTK) substrate) in FP dilution buffer (part #P2839, Invitrogen, Carlsbad, Calif.). After a 30 minute incubation, the fluorescence polarization of each composition in the plate was read on a Tecan Ultra plate reader using a 485 nm excitation filter (20 nm bandpass) and 535 nm emission filters (25 nm bandpass). Data was collected using 10 flashes per well and a 40 μs integr...
example 3
Competition Curve between Labeled Kinase Product Tracer and Unlabeled Kinase Product
[0298] A competition curve to show that the disruption of the antibody-tracer interaction could be monitored by both fluorescence polarization and time-resolved RET from the same sample was performed by incubating serial dilutions (10 μM to 19.5 nM in two-fold dilutions) of an unlabeled phosphotyrosine-containing peptide competitor (ADE(pY)LIPQQS, where pY is a phosphorylated tyrosine, SEQ ID NO:3) in the presence of 10 nM Tb-chelate labeled PY72 antibody and 1 nM labeled PTK labeled tracer, as described above. After a 30 minute incubation, the plate was read on a Tecan Ultra plate reader. Fluorescence polarization was measured using a 485 nm excitation filter (20 nm bandpass) and 535 nm emission filters (25 nm bandpass). Time-resolved RET was measured using a 340 nm excitation filter (35 nm bandpass) and 495 nm (10 nm bandpass) and 520 nm (25 nm bandpass) filters using a 200 μs integration window a...
PUM
| Property | Measurement | Unit |
|---|---|---|
| range of wavelength | aaaaa | aaaaa |
| range of wavelength | aaaaa | aaaaa |
| range of wavelength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com