Xylose isomerase, and encoding gene and use thereof
A technology of xylose isomerase and encoding, applied in the field of enzyme genetic engineering, can solve the problem of insufficient specific enzyme activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0068] The xylose isomerase gene mxylA provided by the present invention is obtained by cloning the xylose isomerase gene xylA (Genebank EU643621) from Sorangium cellulosum 157-2, and then combined with DNA shuffling by error-prone PCR In vitro molecular directed evolution technology introduces mutation points and obtains them through screening. The specific steps are as follows:
[0069] (1) Total DNA was extracted from Sorangium cellulosum 157-2:
[0070] Collect 0.1 g of cells, add 0.75 mL of GTE buffer (25% sucrose, 50 mmol / L Tris-Cl, 100 mmol / L LEDTA, pH 8.0) and appropriate amount of glass beads, vortex, and mix the cells thoroughly. Transfer the bacteria to a 5mL centrifuge tube, add 0.5mL GTE, mix with a vortex shaker, add 150μL of 10% SDS, 15μL of 20mg / mL proteinase K, mix well, and incubate at 37°C for 1h. Invert and mix at regular intervals during this period. Add 0.25mL 5mol / L NaCl, mix well and let stand for 10min, then add 0.25mL CTAB / NaCl (10%CTAB / 0.7mol / L NaCl)...
Embodiment 2
[0087] Enzyme activity assay of evolution enzyme S1:
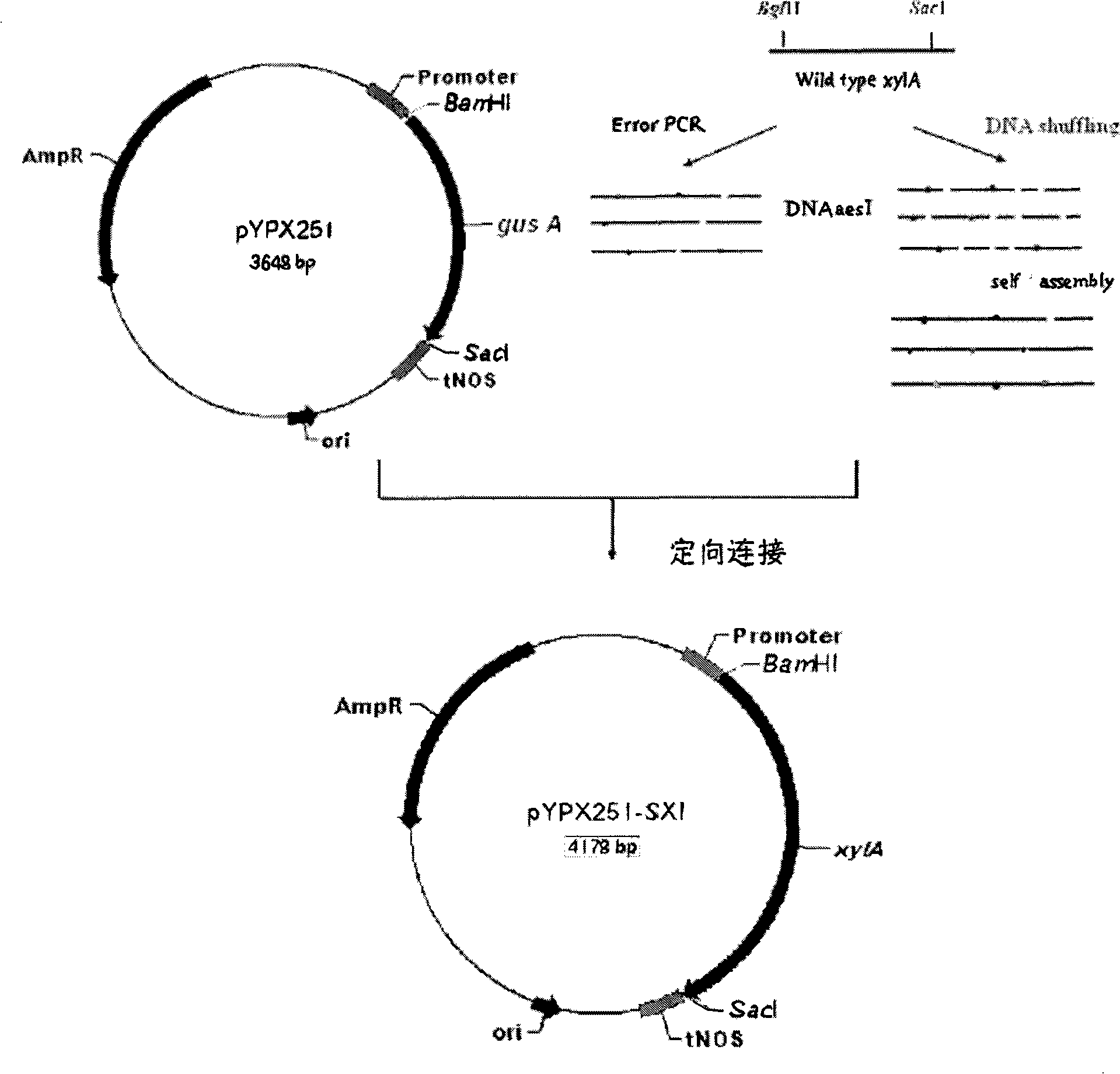
[0088] (1) Using the total DNA of Sorangium cellulosum 157-2 as a template, use primer 1 and primer 2 to amplify the wild-type xylose isomerase gene xylA, digest with BglII and SacI, and adopt a directional ligation strategy Directly ligated with the plasmid vector pYPX251 (GenBank AY178046) treated with the same enzyme digestion to obtain the recombinant plasmid pYPX251-SXI.
[0089] (2) Escherichia coli HB101 was transformed with the recombinant plasmid in step (1) to obtain recombinant strain HR101-SXI.
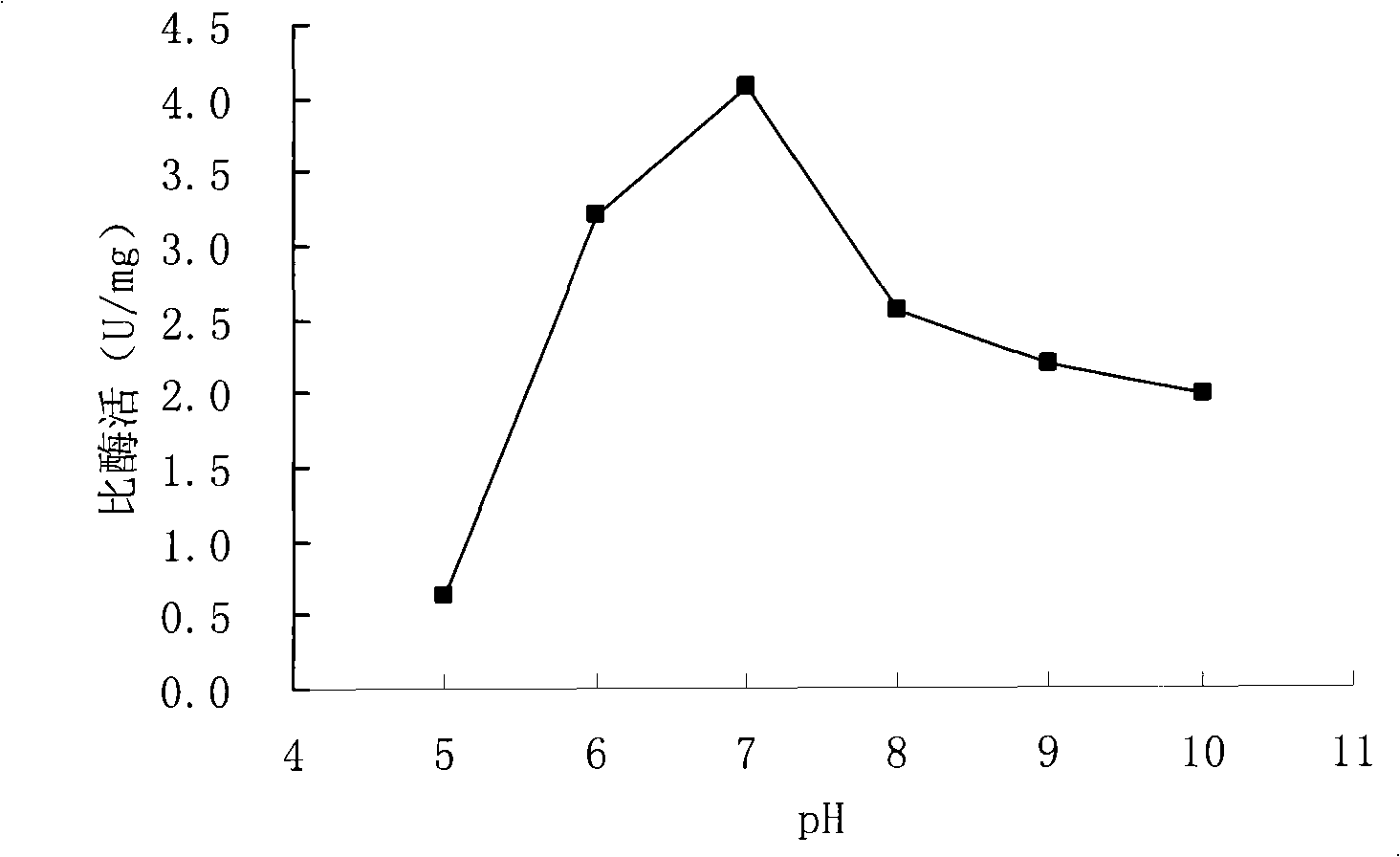
[0090] (3) According to the enzyme activity assay method described in step (11) in Example 1, measure different bacterial strains at different temperatures
[0091] The specific activity of xylose isomerase contained in the source crude enzyme solution is shown in Table 1.
[0092] Table 1 Specific activity of xylose isomerase in different strains at different temperatures
[0093]
[0094] (4) According to the e...
Embodiment 3
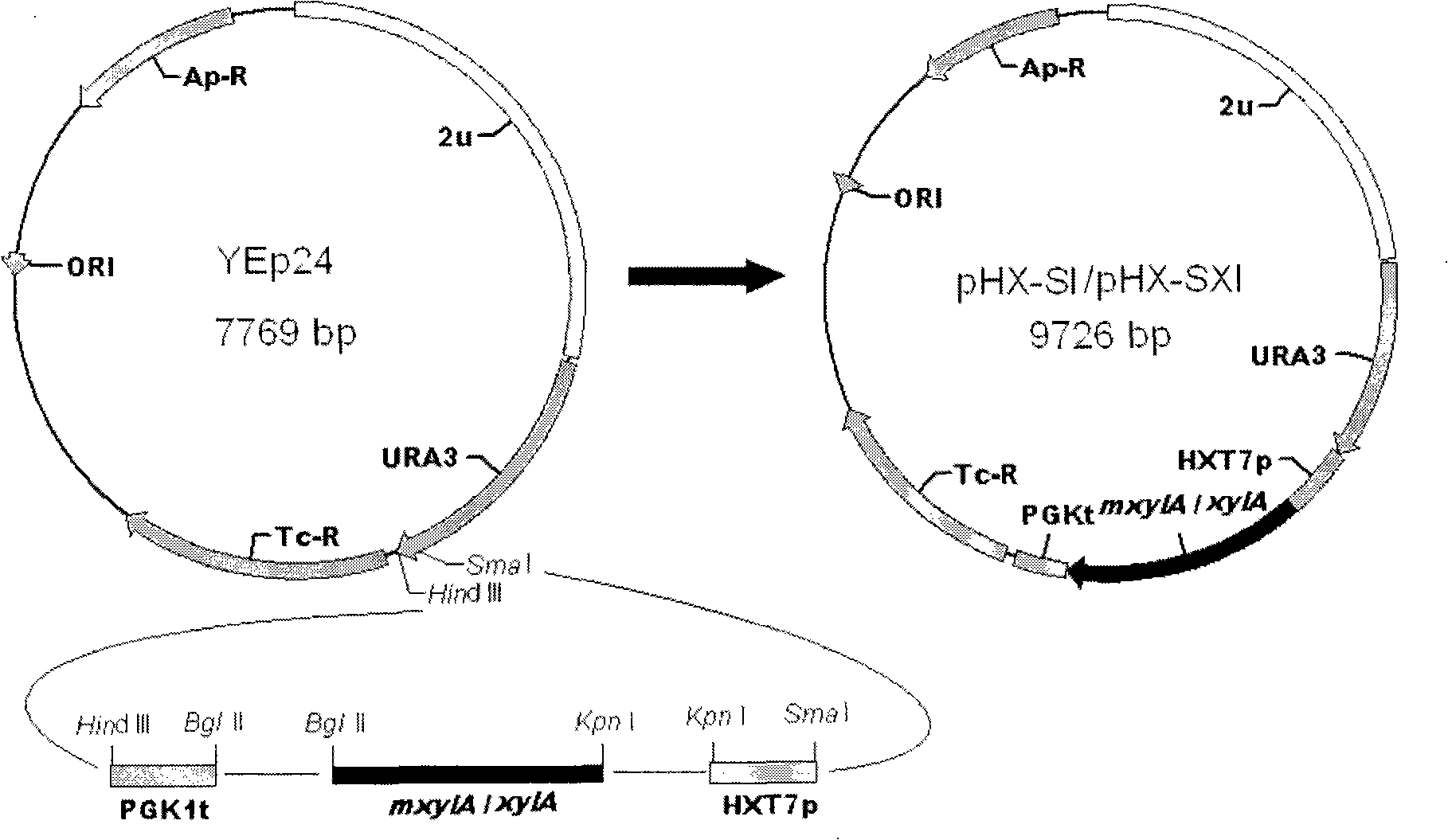
[0096] Example 3 Active Expression of Mutant Xylose Isomerase Gene in Saccharomyces cerevisiae
[0097] (1) Design primers according to the upstream-375bp--1bp sequence of the HXT7 gene in the Saccharomyces cerevisiae genome database:
[0098] Primer 3: 5'-GATC CCCGGG CGGGCCCCTGCGTG-3'
[0099] Primer 4: 5'-TTGC GGTACC TTTTTGATTAAAATTAAAAAAAAC-3'
[0100] The HXT7 promoter (HXT7p) fragment was amplified by PCR using the Saccharomyces cerevisiae chromosome as a template, and the PCR product was double-digested with SmaI and KpnI.
[0101] (2) Design primers according to the nucleotide sequence shown in SEQ NO.1:
[0102] Primer 5: 5'-AGCT GGTACC ATGACCGTCGTGATTGGAAAC-3'
[0103] Primer 6: 5'-TTGC AGATCT TTACCGGATCCACCGATTGAC-3'
[0104] Using pYPX251-S1 as a template, mxylA was amplified by PCR, and the PCR product was double-digested with KpnI and BglII.
[0105] (3) Design primers according to the nucleotide sequence shown in SEQ NO.1:
[0106] Primer 5: 5'-AGCT ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com