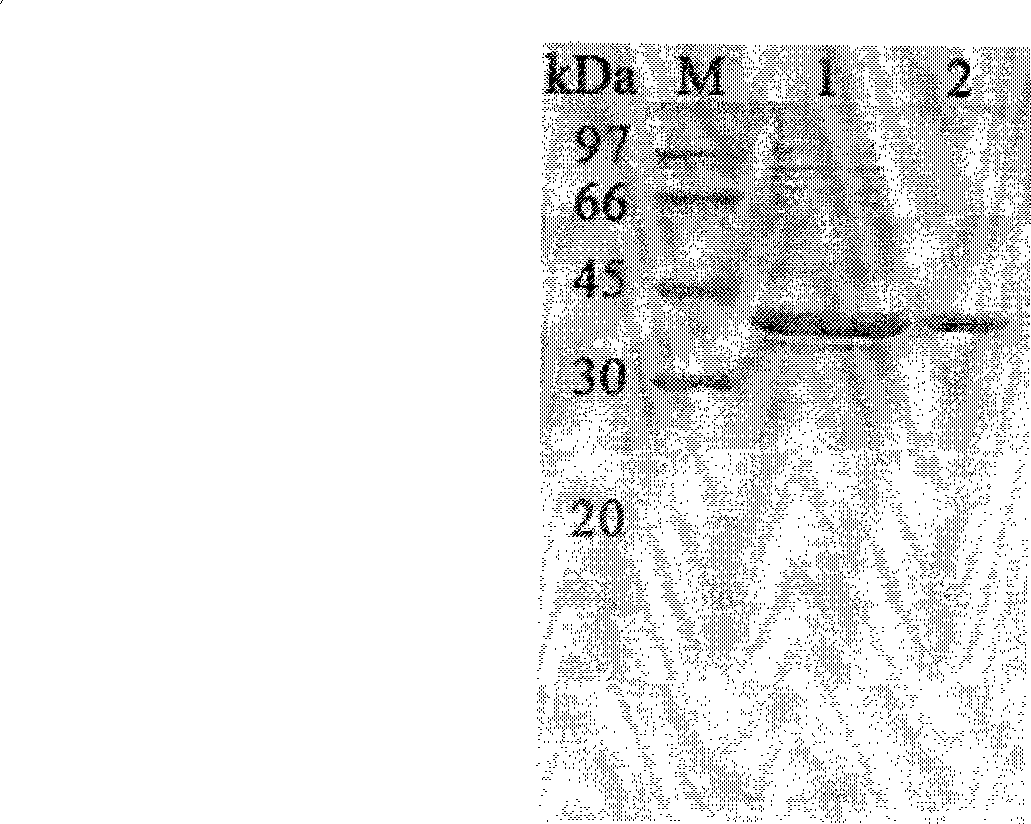
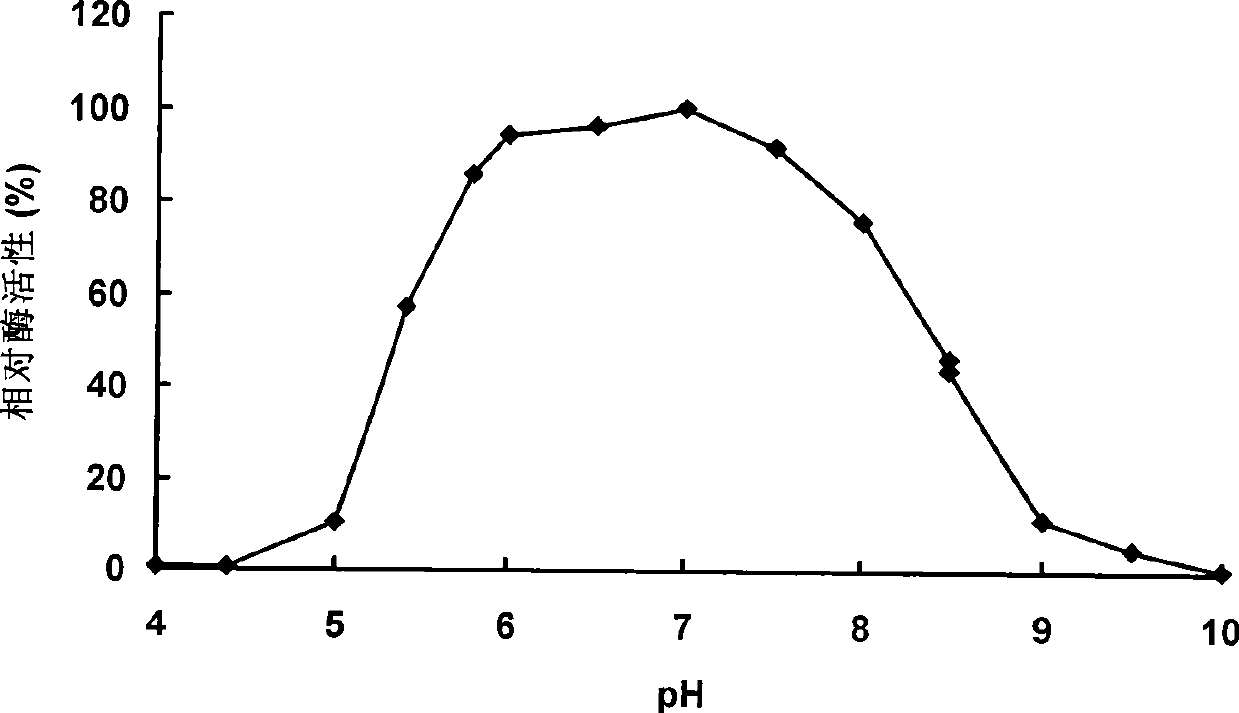
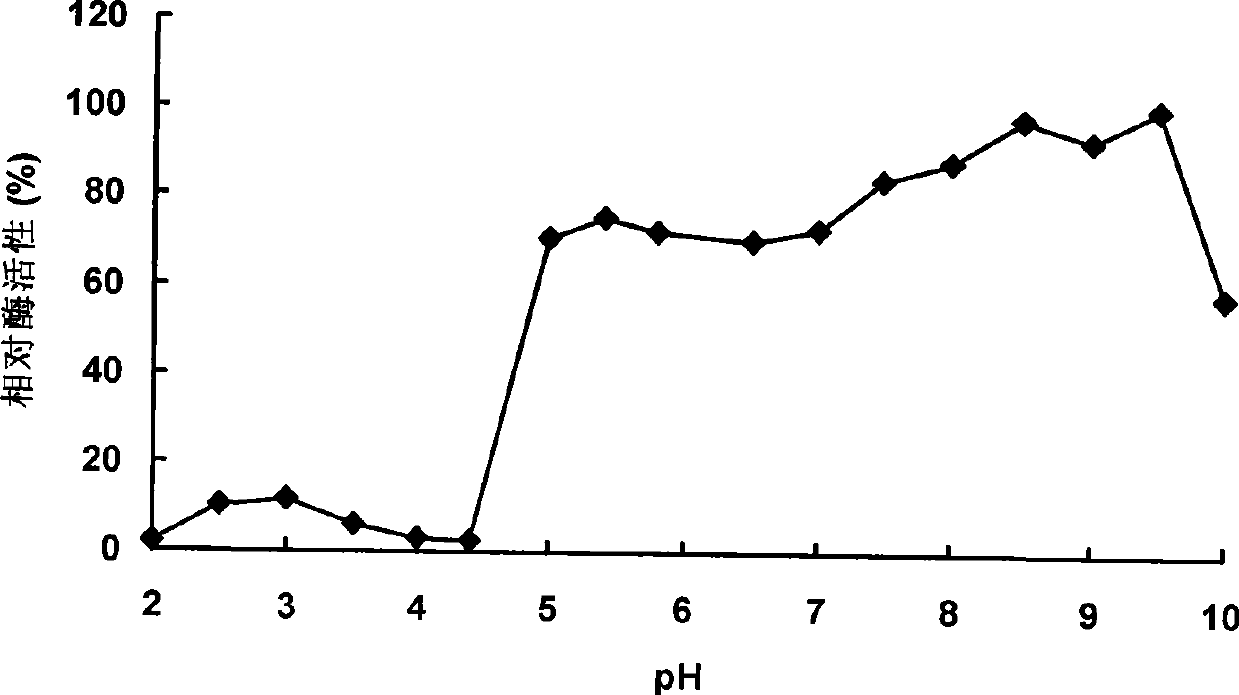
Neutral phytase PHYMJ11 and gene and application thereof
A phytase and neutral technology, applied in the field of genetic engineering, can solve the problems of increasing feed costs, phytate phosphorus cannot be effectively used by animals, and waste of phosphorus sources
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0076] Example 1 Screening and Characterization of Phytase-producing Agrobacterium (Pedobacter sp.MJ11)
[0077] Pedobacter sp.MJ11 is derived from the root soil of Saussurea chinensis in low temperature environment and has low temperature characteristics. Its genomic DNA was extracted, and determined by 16S rDNA sequence, it had the highest sequence similarity with Pedobacter sp.Ellin108, combined with morphological observation and physiological and biochemical characteristics, it was determined that it belonged to the genus Pedobacter. After activation with solid LB medium, transfer to low phosphorus medium (LPM, containing 1 g of casein peptone; 0.45 g of soybean peptone; 4 g of glucose; 0.5 g of glutamic acid; 5 g of sodium chloride; MgCl2 1.7 mM; MgSO4 1.4 mM ; KCl, 0.47mM; CaCl2 0.3mM; dissolved in 900mL 50mM pH7.5 Tris-HCl, dilute to 1000mL, aliquoted, sterilized at 15 lbs for 15min for use.), 30°C constant temperature rotary shaking culture for 24-36h. Take the supern...
Embodiment 2
[0078] Example 2 Cloning of the phytase-encoding gene phyMJ11 from Agrobacterium phytase
[0079] To extract the genomic DNA of Pedobacter sp.MJ11: take the bacterial solution cultured at 37°C for 2 days and centrifuge at 10000rpm for 10min. Take 100 mg of bacterial cells and add 500 μL of sterile water to wash, and centrifuge to get the precipitate. The precipitate was resuspended in 500 μL extract mixture, incubated at 37°C for 60 min, and centrifuged at 10,000 rpm for 10 min to remove the precipitate. The supernatant was extracted sequentially with equal volumes of phenol, phenol:chloroform, and chloroform. Take the upper layer solution and add 0.6-1 times the volume of isopropanol to precipitate at room temperature for 10 minutes. Centrifuge at 12000rpm for 15min. The precipitate was washed with 70% ethanol, centrifuged slightly, dried and dissolved in 30 μL sterile water for later use.
[0080] The degenerate primers were designed and synthesized according to the cons...
Embodiment 3
[0082] The activity analysis method of embodiment 3 phytase
[0083] The enzyme activity assay method is: the enzyme liquid is mixed with 0.1mol / L pH7.0 Tris-HCl containing 0.05% BSA and 0.05% Triton X-100 and added 1mM CaCl 2 Dilute with buffer solution, take 50μL diluted enzyme solution plus 950μL of substrate 1.5mmol / L sodium phytate (prepared with 0.1mol / L pH7.0 Tris-HCl buffer, and add 1mM CaCl 2 ), react at 37°C for 15 min, add 1 mL of 10% TCA to terminate the reaction, add 2 mL of chromogenic solution (10 g of ammonium molybdate tetrahydrate + 32 mL of sulfuric acid + 73.2 g of ferrous sulfate, add water to make up to 1 L). For the control, after adding enzyme solution, first add TCA and mix well, then add substrate. After color development, the OD value was measured at 700 nm, and the enzyme activity was calculated.
[0084] An enzyme activity unit (U) is defined as: under certain conditions, the amount of enzyme required to release 1 nmol of inorganic phosphorus per...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com