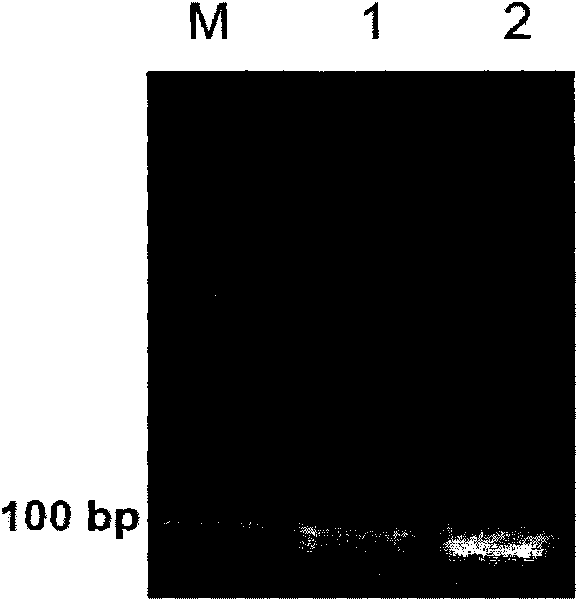High-efficiency separation method for lower molecular multiplex PCR-amplified fragments
A separation method and a technique for amplifying fragments, which are applied in biochemical equipment and methods, recombinant DNA technology, microbial measurement/inspection, etc., can solve the requirements of limited resolution and the inability to meet the detection requirements of multiple PCR small molecule amplified fragments, Complexity and other issues to achieve the effect of improving purity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0069] Example 1 Genomic DNA extracted from crispy rice by CTAB method
[0070] 1) The rice cracker sample containing corn components was ground into powder in liquid nitrogen.
[0071] 2) Weigh 200mg and add it into a 1.5mL Eppendof centrifuge tube, and set up a negative control at the same time.
[0072] 3) Add 1000 μL of CTAB solution, oscillate evenly, incubate at 65° C. for 40 minutes, mix at least 5 times, and centrifuge at 12,000 rpm for 10 minutes at 4° C.
[0073] 4) Carefully pipette the supernatant into another new tube, add an equal volume of Tris-saturated phenol:chloroform:isoamyl alcohol (25:24:1) to the supernatant, shake well, and centrifuge at 12,000 rpm for 15 min at 4°C.
[0074] 5) Repeat step (4) once.
[0075] 6) Carefully pipette the supernatant into another new tube, add an equal volume of chloroform: isoamyl alcohol (24:1) to the supernatant, shake evenly, and centrifuge at 12000 rpm for 15 min at 4°C.
[0076] 7) Carefully pipette the supernatant ...
Embodiment 2
[0082] Embodiment 2 is done PCR detection to the DNA that embodiment 1 extracts
[0083] The DNA extracted in Example 1 was used as a template to amplify the rice crispy genome with the designed primers (as shown in Table 2). The PCR reaction system is as follows (30μL): 10×PCR buffer: 3μL (10×PCR buffer: 100mmol / L Tris-HCl, 500mmol / L KCl, 15mmol / L MgCl 2 ); 2.5mmol / L dNTP: 2.4μL; the primer concentration is 10μM, a total of three pairs of primers, each pair of primers upstream and downstream primers were added 1μL; Taq DNA polymerase: 0.4μL, the extracted DNA was added 1μl as a template, and used wxya 2 O water to make up to 30 μl. The PCR amplification conditions were as follows: pre-denaturation: 94°C for 5 min; denaturation: 94°C for 30 s; annealing: 60°C for 30 s; extension: 72°C for 30 s; final extension: 72°C for 7 min; a total of 35 cycles were set.
[0084] Table 2 Primer design
[0085]
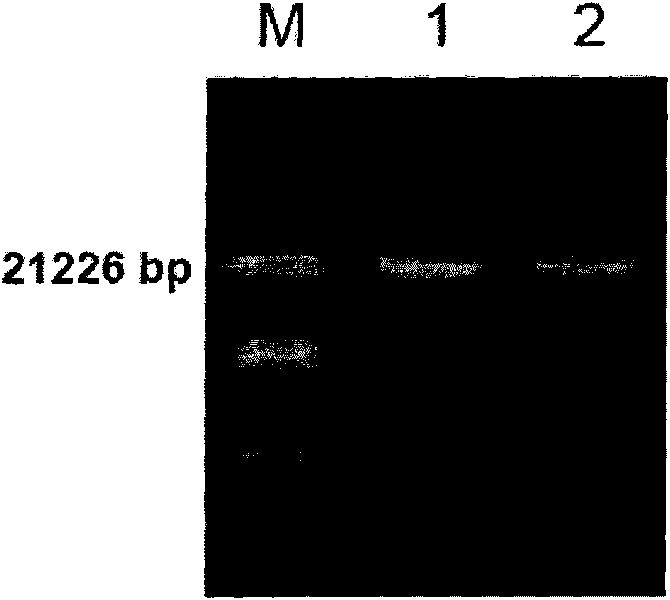
[0086] Such as figure 2 As shown, multiple PCR products were detected ...
Embodiment 3
[0087] Example 3 Purification of Multiplex PCR Small Molecule Amplified Fragments Using Magnetic Bead Purification
[0088] 1) Take the multiplex PCR products in a 1.5ml Eppendorf centrifuge tube. Add an equal volume of magnetic bead binding solution (add 2.0mol / L NaCl, 1.0mol / L MgCl2, 15% PEG 6000 and 15% PEG 8000 to 100mmol / L Tris-Cl, 10mmol / L EDTA solution, the pH value 6.5) and 30uL 1mol / L DTT solution;
[0089] 2) Add 200uL magnetic beads. Before aspirating the magnetic beads, the reagent bottle containing the magnetic beads must be vortexed for 30 seconds to ensure that the magnetic beads are evenly suspended. Pipette repeatedly with a pipette or mix well with a vortex mixer to keep the magnetic beads in a suspended state for 5 minutes.
[0090] 3) Place the centrifuge tube on the magnetic rack and let it stand for 30 seconds. The liquid can only be absorbed when the magnetic beads are completely adsorbed on the side of the centrifuge tube near the magnetic rack. The ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com