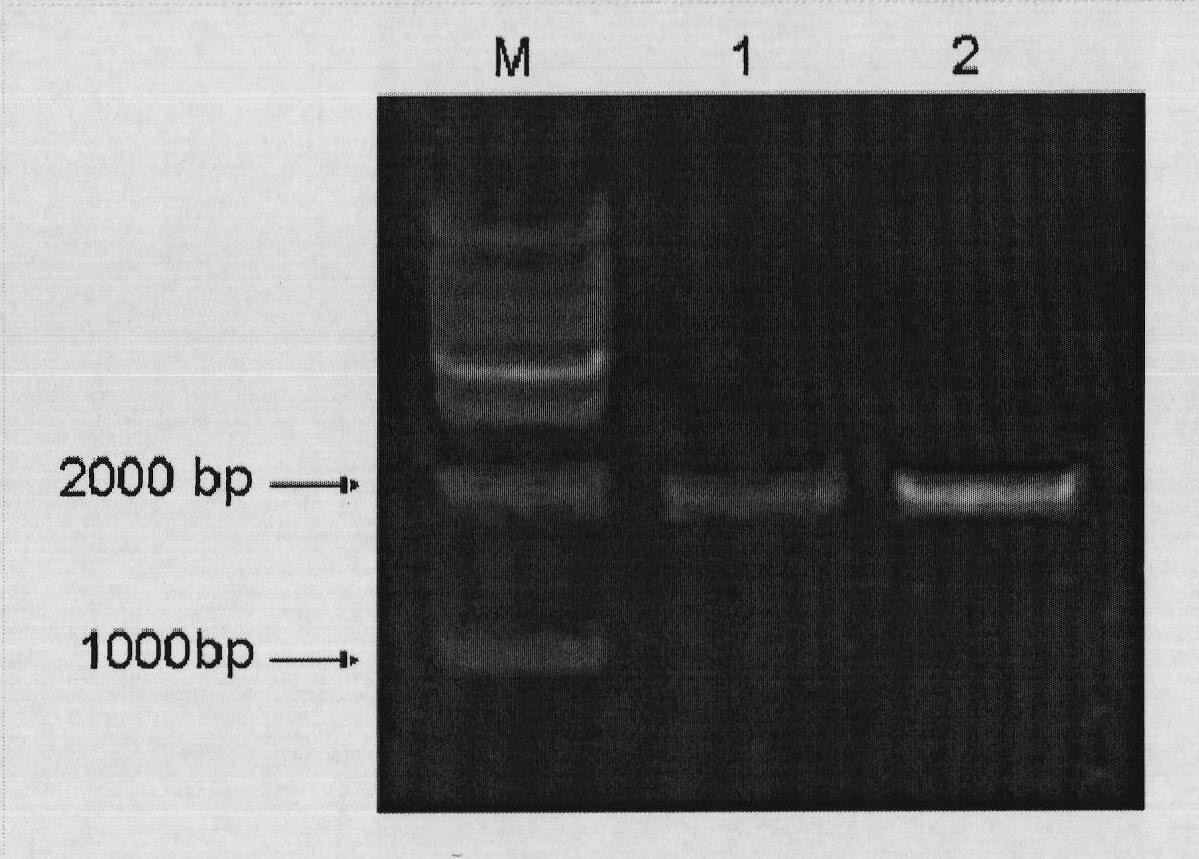
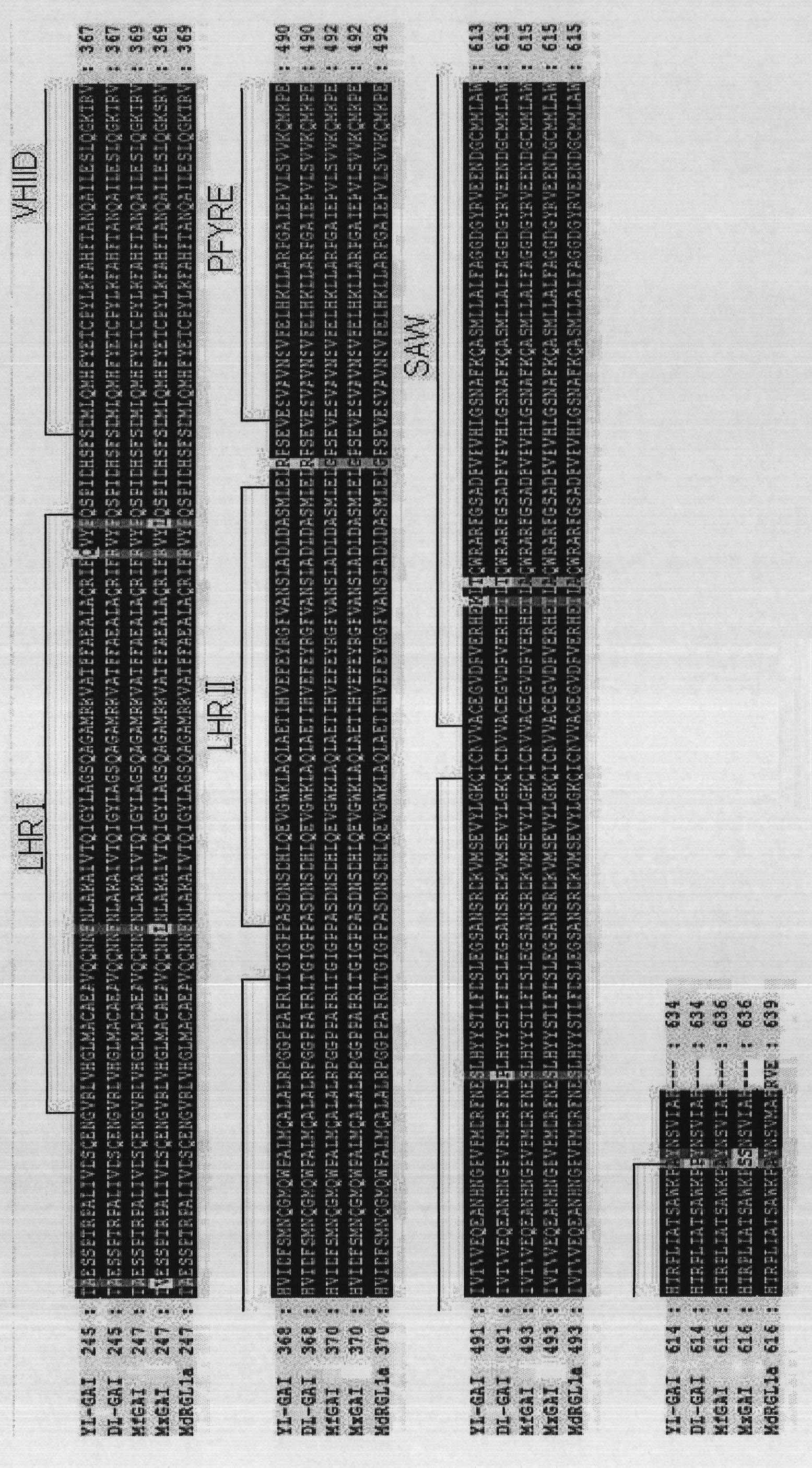
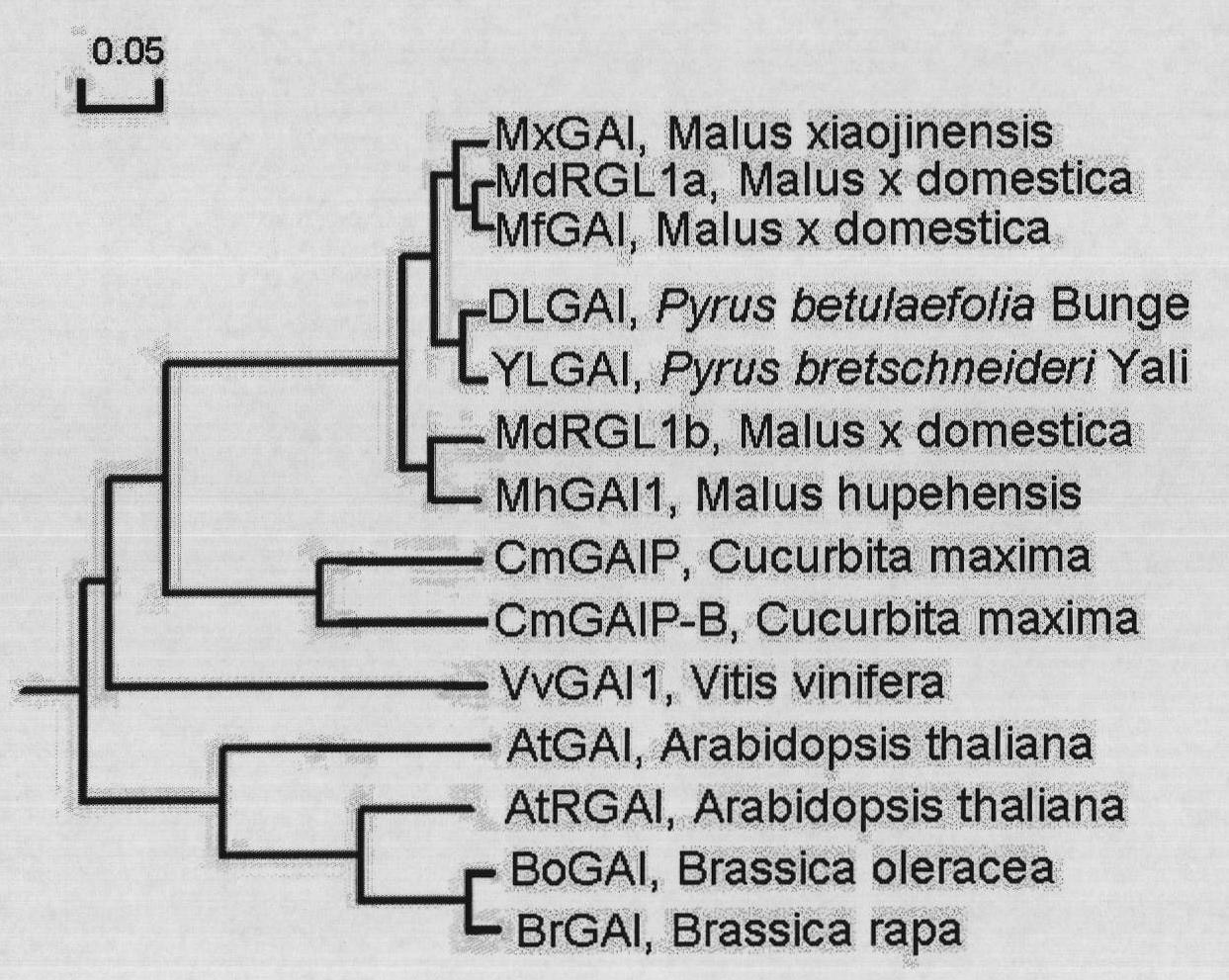
Molecular identification method based on transfer of GAI mRNA molecules between pear rootstock and scion
A technology for molecular identification and duplication of pears, applied in the field of plant molecular biology, can solve problems such as difficulty, heavy workload, and long time, and achieve the effect of rapid method and high accuracy.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1 Test tube micrografting
[0027] Select rootstock Pyrus betulaefolia Bunge and scion 'Yali' (Pyrus bretschneideri 'Yali') tissue culture seedling young leaves, constant light source, light intensity 1500lx, light for 14h, room temperature 23-25°C, relative humidity 85%, 30 ~ 40d subculture once.
[0028]
[0029] plant material medium
[0030]
[0031] ‘Yali’ MS+1.0mg·L -1 6-BA+0.1mg·L -1 IBA
[0032] Du pear MS+0.5mg·L -1 6-BA+0.1mg·L -1 IBA
[0033]
[0034] After liquid nitrogen treatment, they were stored in a -80°C refrigerator for gene cloning. Under sterile conditions, remove the head of the Du pear tissue-cultured seedlings after 30 days of propagation, leave about 1.5 cm long stem section, and cut longitudinally along the top; take the 'Yali' tissue-cultured seedlings wi...
Embodiment 2
[0035] Example 2 Genomic DNA Extraction
[0036] (1) Grind 1g leaves with liquid nitrogen, transfer to a 2.0mL centrifuge tube, add 800μl 2×CTAB (2%CTAB, 10mM Tris-HCl, 1.4M NaCl, 1%PVP), shake slightly to mix, and place at 65°C In the water bath for 20-40min, shake gently several times during the period.
[0037] (2) After cooling down to room temperature, an equal volume of chloroform / isoamyl alcohol (volume ratio 24:1) was added, and centrifuged at room temperature at 12000 rpm for 10 minutes.
[0038] (3) Take the supernatant, add an equal volume of chloroform / isoamyl alcohol (volume ratio 24:1), and centrifuge at 12000 rpm for 10 minutes.
[0039] (4) Add 2 times the volume of absolute ethanol to the supernatant, and precipitate at -20°C for 30 minutes.
[0040] (5) Centrifuge at 12000 rpm for 10 minutes, discard the supernatant, wash the precipitate twice with 75% ethanol, dry the precipitate, and dissolve in an appropriate amount of double distilled water.
[0041] (6)...
Embodiment 3
[0044] Example 3 GAI genome full-length cloning
[0045] Primers were designed based on the conserved regions of the GAI genes of plants such as Arabidopsis thaliana, squash, and apple 'Fuji' published in GenBank.
[0046] The primers used to amplify the GAI gene are as follows:
[0047] Upstream primer: 5'-TTGATTTCCGAGCCCTACCC-3',
[0048] Downstream primer: 5'-AACTCGGTCATCGCTCACTGA-3'.
[0049] The above primers were synthesized by Shanghai Sangon Bioengineering Co., Ltd.
[0050] PCR reaction system: Prime STAR TM HS DNA polymerase (Takara company, DR010S), Takara LA PCR TM in vitro Cloning Kit (Takara, DRR015)
[0051]
[0052] 5 x Prime STARs TM Buffer 10μl
[0053] dNTP mixture (2.5mM each) 4μl
[0054] Primer F / R (10μmol / L) 1μl
[0055] Templete 1μl
[0056] Prime STAR TM HS DNA aggregate (2.5U / μl) 0.5μl
[0057] Double distilled water 32.5μl
[0058] ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com