Penicillium source chitosanase gene and preparation method thereof
A technology of chitosanase and gene, which is applied in the field of genetic engineering and microorganisms, can solve the problems of less research on cloning and expression, and achieve the effect of effective and reliable method, saving time and high activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
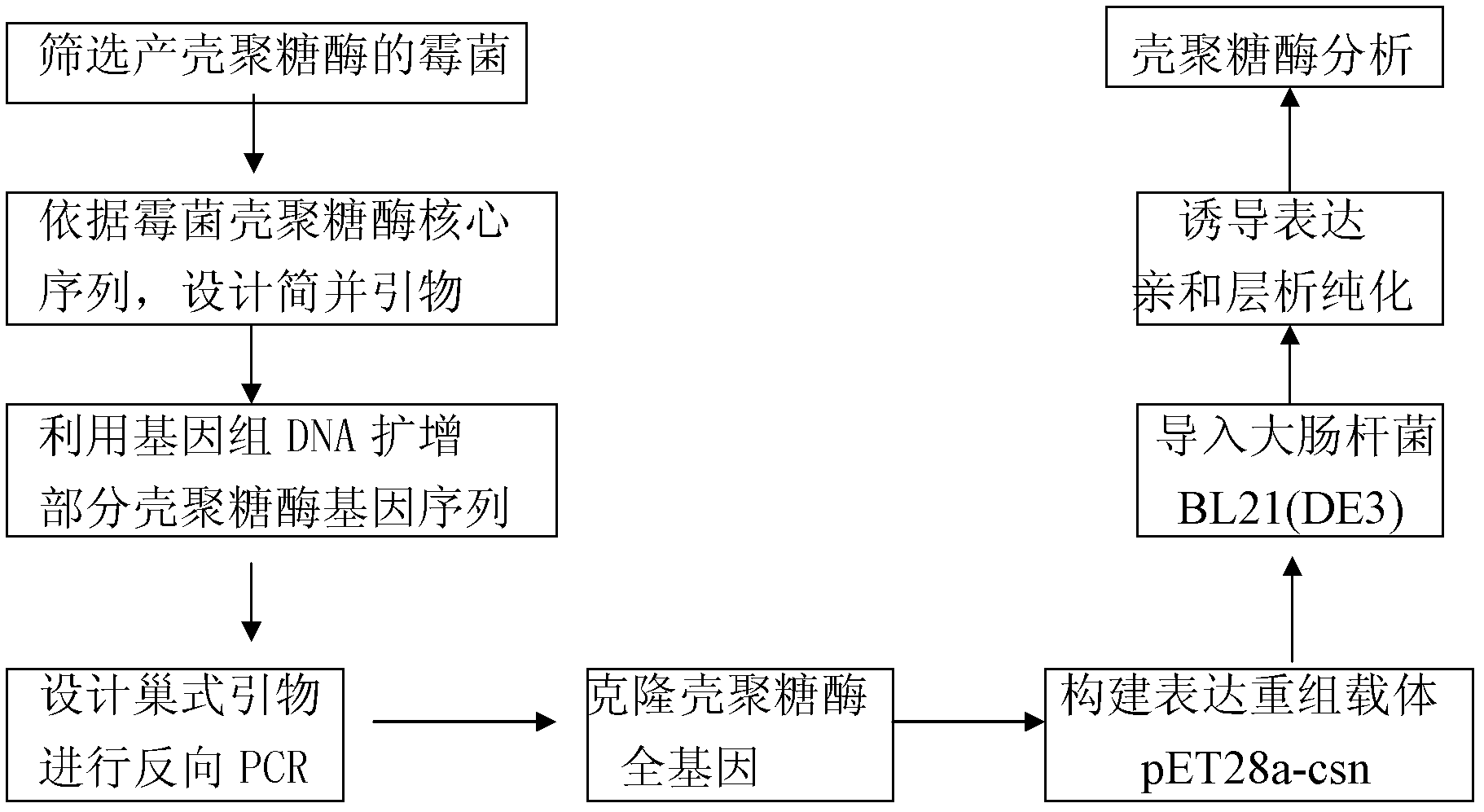
[0023] The acquisition of embodiment 1 penicillium
[0024] Using chitosan as a single carbon source medium (NaNO 3 0.2%, K 2 HPO 4 0.1%, MgSO 4 0.05%, KCl 0.05%, FeSO 4 0.001%, colloidal chitosan 0.5%, pH 5.0) cultured and screened to obtain the common Penicillium sp) that can express and produce chitosanase.
Embodiment 2
[0025] Cloning of embodiment 2 chitosanase gene
[0026] After the Penicillium thallus DNA of Example 1 was extracted, the chitosanase gene was obtained by inverse PCR (I-PCR). Firstly, the primers DFP (5'-GCGGATCCAAYATGGAYATHGAYTGYGA-3') and DRP (5'-GCGAAGCTTRTCDCCCCADATNCCRTA-3') were designed through the chitosanase highly conserved sequence of the fungus, and the genomic DNA of Penicillium was used as a template for PCR. The program was 94 Pre-denaturation at ℃ for 5 min, denaturation at 94 °C for 30 s, annealing at 58 °C for 1 min, constant temperature at 72 °C for 2 min, 30 cycles, and finally extension at 72 °C for 10 min. The obtained fragments were sequenced, and then nested primers FP1 (5-CCA GAG CAC GTT GGC ATC AA-3), FP2 (5'-GGT CTG CAA CAA CAA GCT CAT C-3') and RP1 were further designed according to the obtained sequences (5-ACC ATA GTC GGA CTT GAC CT-3), RP2 (5'-GAG TCG ATG CCG TCT TGA TC-3'). Then use Pst I, BamHI, EcoRI and HindIII to digest genomic DNA respe...
Embodiment 3
[0027]According to the chitosanase full-length coding sequence (SEQ ID NO.1) obtained by gene annotation in the examples, primers capable of amplifying the complete coding reading frame were designed to construct an expression vector. Penicillium DNA was used as a template to amplify the full-length chitosanase gene. Recombined into the expression vector pET28a under the premise of ensuring the correct reading frame, and then transformed the recombinant vector into Escherichia coli DH5α competent cells (the transformation method was CaCl 2 method or electrotransformation method), and antibiotic marker method was used to screen (in this case, kanamycin resistance) positive recombinant bacteria pET28a-csn / DH5α. Pick a single colony of engineered bacteria pET28a-csn / DH5α, shake and culture in 5ml LB medium containing 30μg / ml kanamycin overnight at 37°C, extract the recombinant plasmid and transform it into Escherichia coli BL21(DE3) competent cells , Screening (in this case, kan...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com