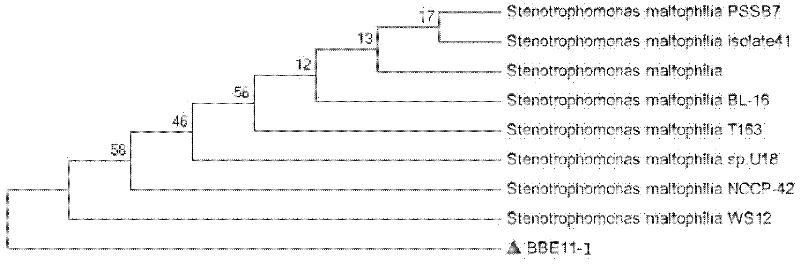
Stenotrophomonas maltophilia for generating keratinase and application of stenotrophomonas maltophilia
A keratinase, monomonas technology, applied in applications, bacteria, microorganism-based methods, etc., can solve problems such as wool texture damage, environmental pollution, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Embodiment 1 produces the screening method of keratinase bacterial strain
[0022] 1. The sampling site is a local poultry farm. Add the soil or sludge samples obtained from the sampling into a triangular flask filled with 20mL of sterilized water and glass beads, mix them on a shaker at 37°C, and crush them to make a bacterial suspension. liquid;
[0023] 2. Add the bacterial suspension to the primary screening medium, 37°C, 200 rpm, until the feathers are degraded, and pass for three times.
[0024] Primary screening medium: Feather 10, K 2 HPO 4 , 1.4, KH 2 PO 4 0.7, NaCl0.5, MgSO 4 ·7H 2 O 0.1, pH7.0-7.2. (g / l)
[0025] 3. Aspirate the supernatant for gradient dilution, take 10 -4 、10 -6 、10 -6 3 dilutions spread skim milk plates. Incubate upside down at 37°C for 48 hours after coating;
[0026] 4. Select the spots with larger transparent circles and plant them in the skimmed milk medium.
[0027] Six strains could be screened out from the morphologi...
Embodiment 2
[0031] The preliminary fermentation condition determination of embodiment 2 producing keratinase bacterial strain
[0032] Different temperature, pH, nitrogen source, carbon source and inorganic salt were selected to preliminarily determine the fermentation conditions of the screened bacteria. The fermentation time was set at 40h. The results are shown in Table 1.
[0033] Enzyme activity assay: draw 1mL of appropriately diluted enzyme solution, add 1mL of 0.05mol / LTtis-Hcl buffer solution (pH7.5) and 5mg of substrate (feather meal), incubate at 40°C for 60min, add 2mL of 4M TCA solution to terminate the reaction, After standing for 20min, the insoluble matter was filtered off. Centrifuge for 5min, draw 0.5mL supernatant and transfer to a new test tube, then add 0.5mL Folinol reagent and 2mL 0.5M Na 2 CO 3 After developing the color at 40°C for 20 minutes, the absorbance value was detected at 660 nm, and each increase of 0.01 in the absorbance value was defined as one enzyme...
Embodiment 3
[0039] Embodiment 3: keratinase-producing bacteria are applied to feed processing
[0040] The screened bacterial strain is inoculated in the single feather inorganic salt medium with feather content of 20g / L, after 48 hours of fermentation, the fermentation supernatant is taken to detect the free amino acid species and content (Table 2), and the fermentation results are shown in figure 2 .
[0041] Determination of preliminary fermentation conditions of table 2 keratinase-producing strains
[0042]
[0043]
[0044] After amino acid analysis, 17 kinds of amino acids can be detected, including rare amino acids that cannot be synthesized by animals themselves. Adding the fermentation product to poultry feed can replace soybeans to meet the feed used by poultry.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com