HPV16L1 polynucleotide sequence and expression vector, host cell and application thereof
A HPV16L1, polynucleotide technology, applied in the direction of application, introduction of foreign genetic material using vectors, chemical instruments and methods, etc., can solve problems such as acceptance by the general public
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
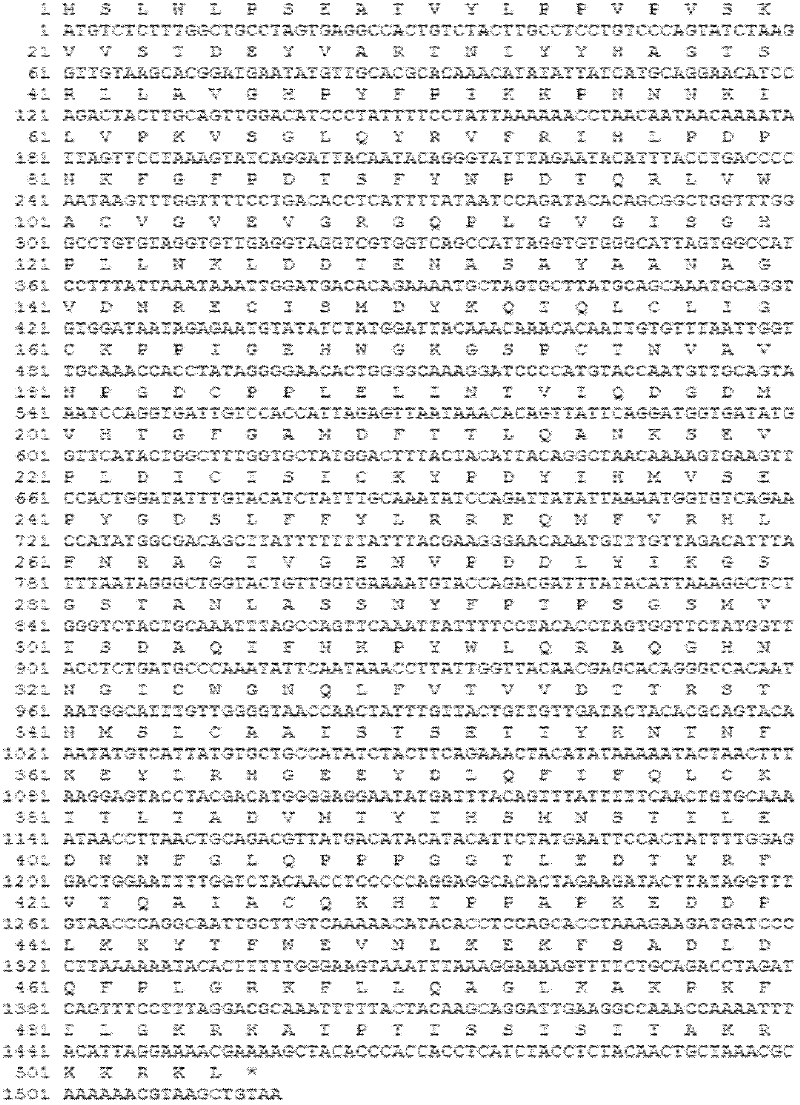
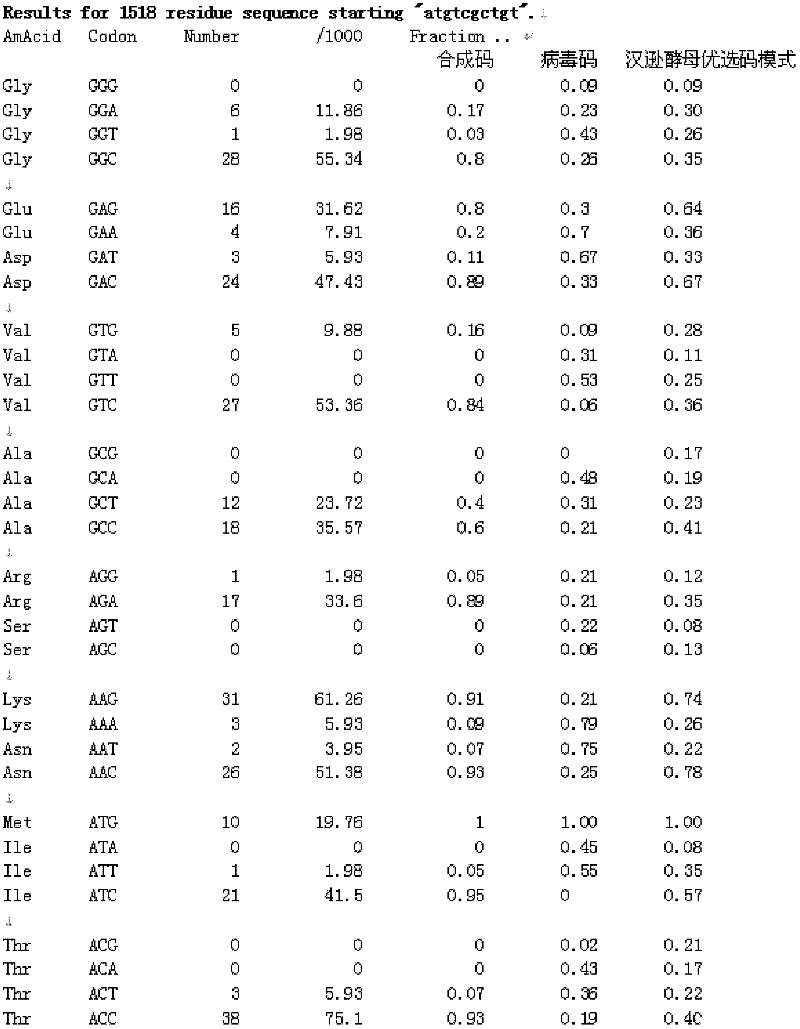
[0074] Example 1: L1 gene sequence optimization
[0075] Convert the full-length 506 amino acids of the HPV16L1 amino acid sequence into the nucleotide sequence of Hansenula's preferred codon pattern, and the codon with Hansenula's preference for the first codon in the preferred demerging codons is mainly used in the Hansenula codon. Yeast prefers the first and second codons, and in a few cases, the third codon is used to complete local adjustments. DNAMAN software is used to avoid excessively long complementary sequences and repetitive sequences, and RNA structure prediction is used to avoid excessively long and complex hairpin structures. , to exclude strong secondary structures and increase the internal stability of specific molecules. The nucleotide sequence of the gene also avoids sequences that may adversely affect gene transcription and translation efficiency, such as intron splicing sequences, transcription termination sequences, internal promoter sequences (TATA) and ...
Embodiment 2
[0077] Embodiment 2: HPV16L1 gene and HPV16L1 mutant gene expression vector construction
[0078] The DNA sequence shown in Sequence 2 in the Sequence Listing was synthesized by a fully artificial method, and recognition sites for restriction endonucleases EcoR I and BamH I were added to both ends of the sequence. The synthesized DNA sequence was digested with EcoR I and BamH I, and the digested fragment was recovered by electrophoresis, and stored at -20°C for future use.
[0079] Using the genomic DNA of Hansenula polymorpha CGMCC 2.2497 as a template, clone the 1.5kb methanol oxidase gene (MOX) promoter, the 350bp methanol oxidase gene (MOX) terminator, and the 1.0kb self-replicating sequence HARS; and from YIp5 (GeneBank Accession NO.L09157) plasmid cloned the 1.1kb Saccharomyces cerevisiae uracil gene ScURA3; after connecting the above four parts, it was inserted into the multiple cloning site of the pBluescrip II plasmid to construct the shuttle vector pMPT-02.
[0080]...
Embodiment 3
[0085] Example 3: Electrotransformation of Hansenula Competent Host Cells with Expression Vectors
[0086] The vector pMPT-HPV16L1 was digested with Sac I to linearize the vector. The linearized vector was transformed into Hansenula polymorpha (Hansenula polymorpha) CGMCC NO.1218 by electroporation method (LN-101 gene pulse introduction instrument produced by Tianjin University of Technology, 1.5kV, 50μF, 200Ω, 3-5mSec). Transformants were selected on a medium plate containing 1.34g / 100ml YNB (yeast nitrogen base medium), 2g / 100ml glucose.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com