Sugar cane sucrose transport protein ShSUT3 and application of coding gene thereof
A technology of sucrose transporter and encoding gene, applied in the field of sucrose transporter ShSUT3 and its encoding gene and application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
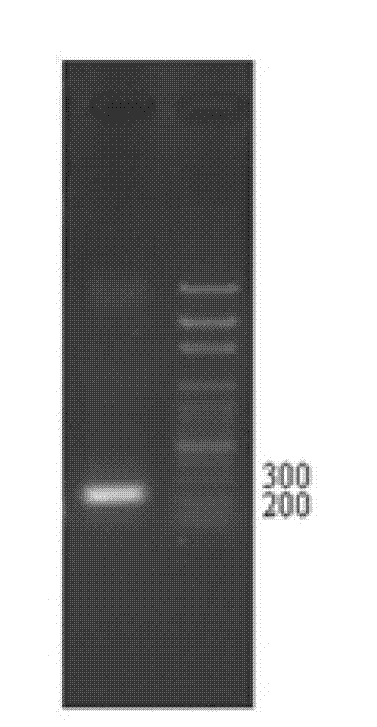
[0036] Example 1 Cloning and analysis of the full-length coding region of the ShSUT3 gene
[0037] 1. Extraction of total RNA
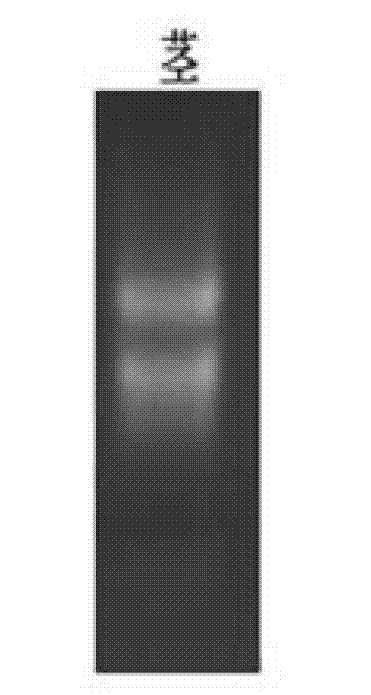
[0038] Take 0.1 g of fresh sugarcane stems with 5-7 nodes (counting downward from the growth point), cut them into flakes, and grind them into powder in liquid nitrogen; add 1 mL Trizol (Gibco, Japan), and extract total RNA according to the kit instructions. Carry out 1.2% agarose gel electrophoresis detection, the result is as follows figure 1 shown. The extracted RNA has two obvious electrophoresis bands, which are 28S RNA and 18S RNA from top to bottom, indicating that the total RNA with higher purity and integrity has been obtained.
[0039] 2. Synthesis of the first strand of cDNA
[0040] Mix 5 μg of sugarcane total RNA with 1 μL (10 pmol / L) of reverse transcription primer (oligo-dT linker primer), heat at 70 °C for 5 min, place it on ice immediately, and then add 5× buffer, 2.5 mmol / L dNTP mixture, Ribonuclease Inhibitor, M-MLV reverse tran...
Embodiment 2
[0064] Example 2. Bioinformatics analysis of ShSUT3 and its encoded protein
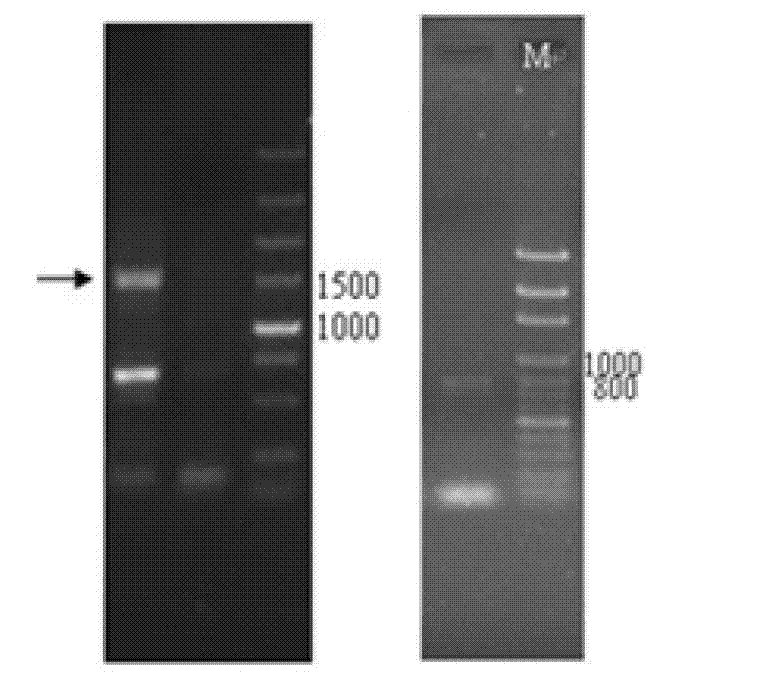
[0065] Utilize DNAMAN and OMIGA software to carry out bioinformatics analysis to the full-length cDNA sequence of ShSUT3 obtained in embodiment 1, ShSUT3 this sequence full-length 1605bp, wherein from the 1st position of 5' end to 1605th position is a complete ORF, coded by 534 A protein composed of amino acid residues. The schematic diagram of the structure of ShSUT3 is shown in Figure 5 shown. The domain of ShSUT3 was analyzed with the online Blast tool, and the results showed that the protein belonged to the GPH sucrose superfamily, indicating that the protein was a member of the SUT family. The hydrophobicity of the protein was predicted by the membrane protein online prediction tool TopPred2 ( Image 6 ) and is a 12-transmembrane protein ( Figure 7 ), conforming to the structure of the sucrose transporter family, indicating that it belongs to the sucrose transporter family.
Embodiment 3
[0066] Example 3. Functional analysis of ShSUT3 in yeast mutants
[0067] The ShSUT3 gene amplified in Example 1 was amplified by primers containing SpeI and XhoI restriction sites (primer sequence F: 5'-GGACTAGT ATTTGCCTGTCCCTATCGTCCTCT-3', R: 5'-GGGAATTCCGGCAGACACCAAGAATCAGACTC-3'), and the amplified The amplified target fragment was recovered and purified, connected to the pMD-19T simple (TaKaRa Code: D104A) vector, the ligated product was transformed into Escherichia coli JM109 competent cells, and the positive clones were screened for bacterial liquid PCR identification, and the plasmids of the positive clones were extracted Perform sequencing. Sequencing results showed that the amplified fragment had the nucleotide sequence of the coding region in sequence 2. The plasmids in the clones with correct sequencing results were extracted, digested with SpeI and XhoI, and inserted between the SpeI and XhoI restriction sites of the yeast expression vector PDR196 that had been d...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com