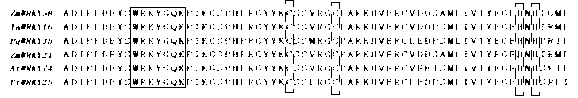Corn WRKY transcription factor ZmWRKY58, and coding gene and application thereof
A technology of transcription factors and coding genes, which is applied in the fields of genetic engineering and crop genetics and breeding, and can solve the problems of few research reports on other species
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Example 1 Acquisition of Maize WRKY Family Gene ZmWRKY58
[0036] 1. Stress treatment: the plant material is corn B73 inbred line, selected seeds with uniform size and full grains are sown in flower pots with sandy soil, and distilled water is sprayed timely to keep the soil moist. When the seedlings grew to two leaves and one heart, the normal irrigated plants were used as controls, and the stress treatments of 16% PEG-6000 and 200mmol / L NaCl were carried out respectively, and the treatment time was 12, 24, and 48 hours respectively. The scissors cut the leaves of the same part of the maize seedlings in the test group and the control group respectively, and three samples were taken at each treatment stage. After sampling, the samples were immediately frozen in liquid nitrogen and stored in a -80°C refrigerator.
[0037]2. RNA extraction: The corn material obtained above was ground by adding liquid nitrogen, and then quickly transferred to a 1.5 ml centrifuge tube (pre...
Embodiment 2
[0043] Example 2 Sequence Homology and Homology Analysis of Maize ZmWRKY58
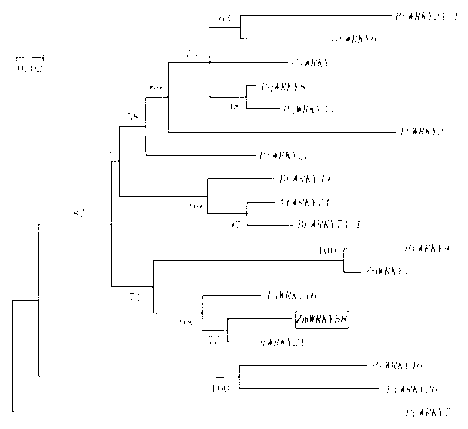
[0044] According to the sequence sequencing results, the sequence comparison was performed in the NCBI database, and it was found that the cloned gene sequence had the closest homology relationship with the WRKY family transcription factors. Comparing the protein sequence of this transcription factor with other members of the WRKY transcription factor family, the structure type of its zinc finger is analyzed as C2H2 type. According to the structural characteristics of the DNA binding domain, the ZmWRKY58 protein belongs to the class II WRKY transcription factor family (such as figure 1 ). In order to further analyze the phylogenetic relationship between ZmWRKY58 and other WRKY transcription factors, the phylogenetic relationship between WRKY proteins and ZmWRKY58 in different plants was analyzed (such as figure 2 ).
Embodiment 3
[0045] Example 3 Subcellular localization of ZmWRKY58
[0046] Specific primers were designed with Primer Premier 5.0 software, and the recognition sequences of restriction endonucleases NcoI and SpeI and corresponding protective bases were added to the 5' and 3' ends of the primers, respectively, and handed over to Sangon Biotech. The target fragment was amplified with the correctly sequenced recombinant plasmid P-ZmWRKY58 as a template. The primer sequences are as follows:
[0047] RH-F 5’- ATATCCATGGTAGATGAGGAAGTGGAG-3’
[0048] RH-R 5'-GGACTAGTCACCTGTGCTGCTGCTGC-3'
[0049] The amplified PCR product was recovered and connected to the pMD18-T vector, transformed into Escherichia coli competent by heat shock, a single colony was picked, the plasmid was extracted, and verified by NcoI and SpeI digestion, and the verified correct plasmid was sent for sequencing.
[0050] The above-mentioned recombinant plasmid and pCAMBIA1302 vector were digested with NcoI and SpeI, the tar...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com