Preparation method of PCV2 (Porcine Circovirus2)-D
A technology for porcine circovirus and attenuated vaccine, which is applied in biochemical equipment and methods, antiviral agents, viruses/phages, etc., and can solve problems such as weak and difficult viruses
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Embodiment 1 pET30a-PCV2 whole gene, the construction of exchange recombinant plasmid
[0042] 2.1 Extraction of PCV2 DNA: extract according to the conventional method of phenol chloroform.
[0043] 2.2 PCV2 whole gene sequence amplification
[0044] 2.2.1 Primer design and synthesis
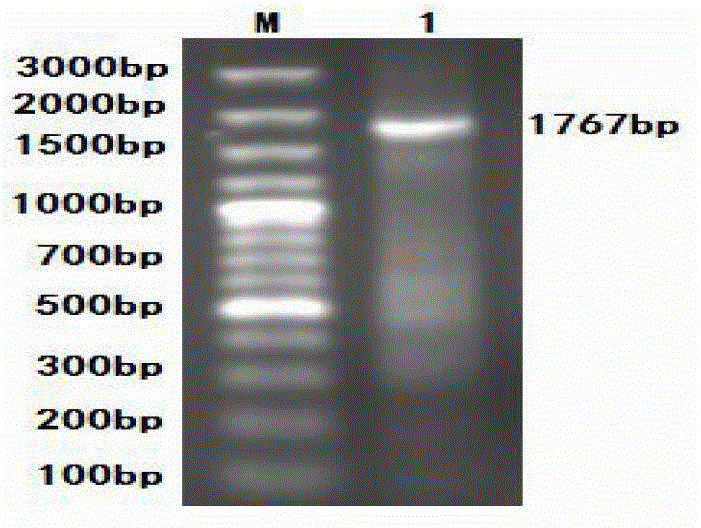
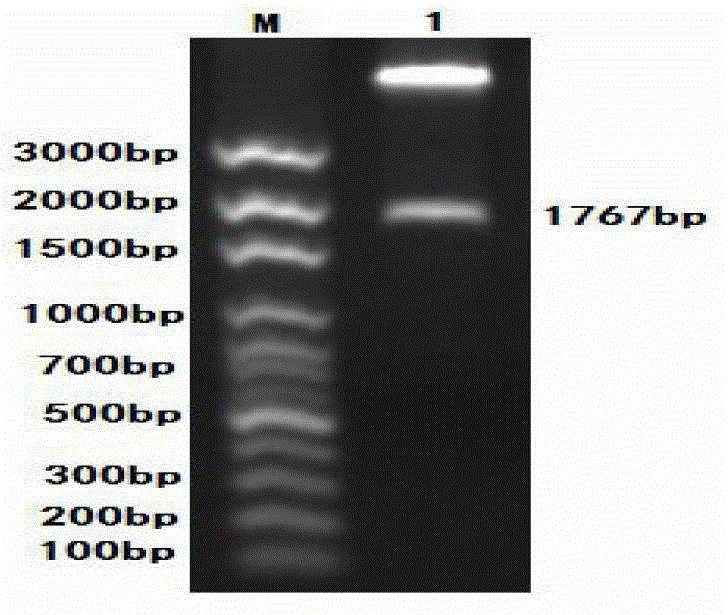
[0045] Design a pair of primers with reference to the sequence on Genbank, marked as PCV-F / R, the specific sequence is as follows, the italics are the enzyme cutting sites, and the full length of the amplified fragment is expected to be 1767bp. Primers were synthesized by Shanghai Jierui Bioengineering Co., Ltd.
[0046] PCV-F: AGAATTCAACCTTAACCTTTCT
[0047] PCV-R: AGAATTCTGGCCCTGCTCC
[0048] 2.2.2 PCR amplification
[0049] PCR reaction system: 2 μL of DNA template extracted from disease materials, 10×Buffer (Mg 2+ free) 2.5 μL, MgSO 4 2 μL, 2 mmol / L dNTP 2.5 μL, 10 μmol / L PCV-F and R primers 1 μL each, KOD-Plus enzyme 1 μL, ddH 2 O 13 μL, total volume 25 μL; mix on ice.
[00...
Embodiment 2
[0077] The construction of embodiment 2PCV2 and PCV2-D strain
[0078] 2.6 Digestion of pET30a-PCV2 and pET30a-PCV2-D plasmids
[0079] Digest pET30a-PCV2 plasmid: 20 μL pET30a-PCV2 plasmid, 3 μL 10×buffer for EcoR I, 3 μL EcoR I, 4 μL ddH 2 O, the total reaction volume is 30 μL; 37 ° C water bath for 2 ~ 3 h.
[0080] Digestion of pET30a-PCV2-D plasmid: 20 μL of pET30a-PCV2-D plasmid, 3 μL of 10×buffer for EcoR I, 3 μL of EcoR I, 4 μL of ddH 2 O, the total reaction volume is 30 μL; 37 ° C water bath for 2 ~ 3 h.
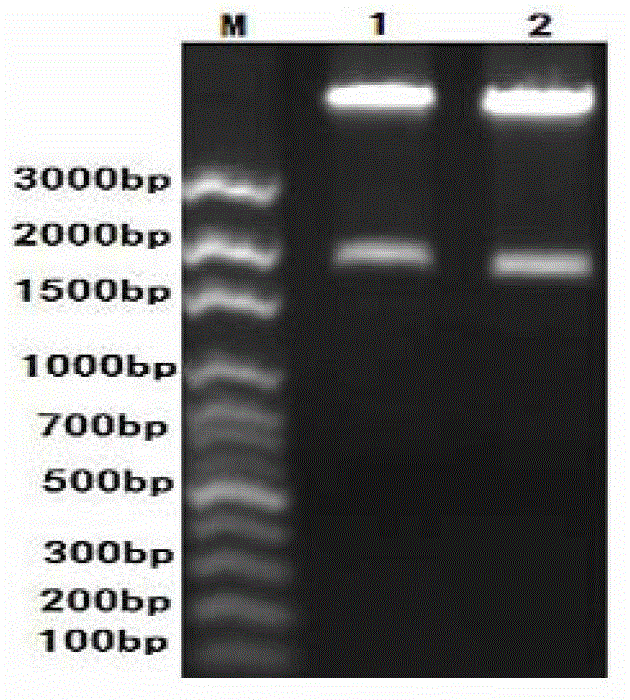
[0081] Results: After the complete digestion was identified by 1% agarose gel electrophoresis, the whole PCV2 gene and PCV2-D gene fragments were recovered with a small DNA gel recovery kit. For enzyme digestion results, see image 3 , indicating that the size of the PCV2 gene fragment before and after transformation was consistent.
[0082] 2.7 Ligation circularization and recovery in vitro
[0083] The linear PCV2 full gene and PCV2-D gene fragments recovere...
Embodiment 3
[0087] Example 3 Transfection of PK-15 cells and the detection of replication and proliferation
[0088] 2.8 Cell transfection test
[0089] (1) Take out a bottle of pk15 cells in good growth state, transfer them to a 24-well plate, and inoculate about 1.25×10 cells per well 5 Put the cells in 500 μL DMEM complete medium without antibiotics at 37°C CO 2 cultured in an incubator. Make sure that the adherent cells in each well grow to 90%-95% before transfection.
[0090] (2) Circularized DNA and transfection reagent dilution
[0091] Put 0.6ug of the circularized PCV2 and PCV2-D plasmids obtained in step 2.7 into 50 μL DMEM without antibiotics and serum.
[0092] Take the transfection reagent Lipofectamine TM 2000 2.4 μL into 100 μL of MEM without antibiotics and serum, shake gently to mix, and incubate at room temperature for 5 minutes.
[0093] (3) Mix the diluted circularized DNA and transfection reagent in equal volumes of 50 μL each, and incubate at room temperature fo...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com