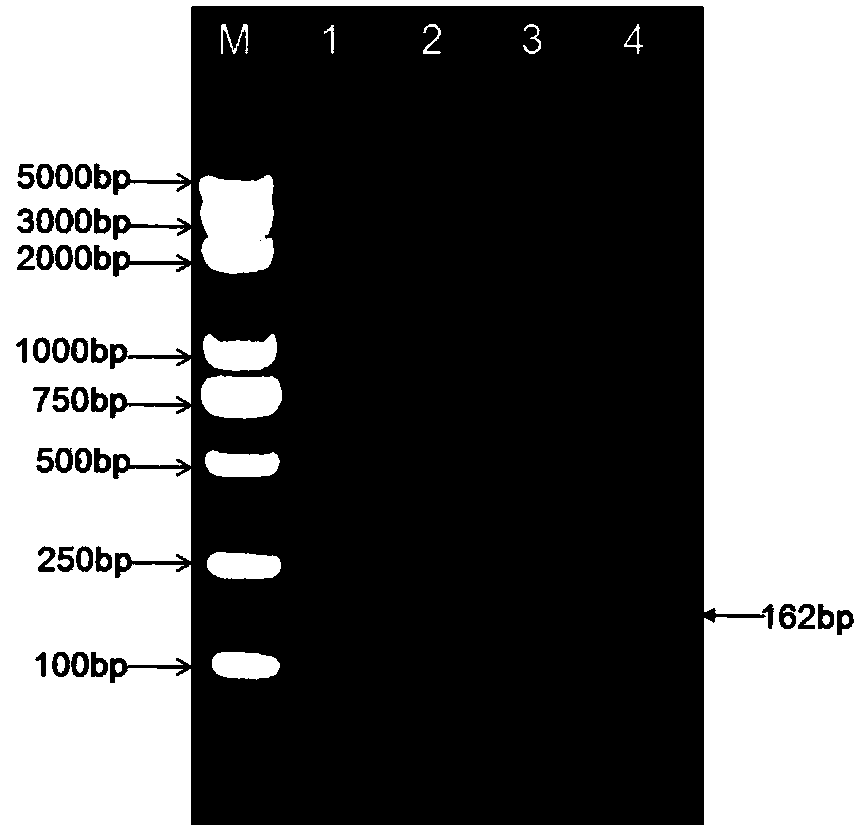
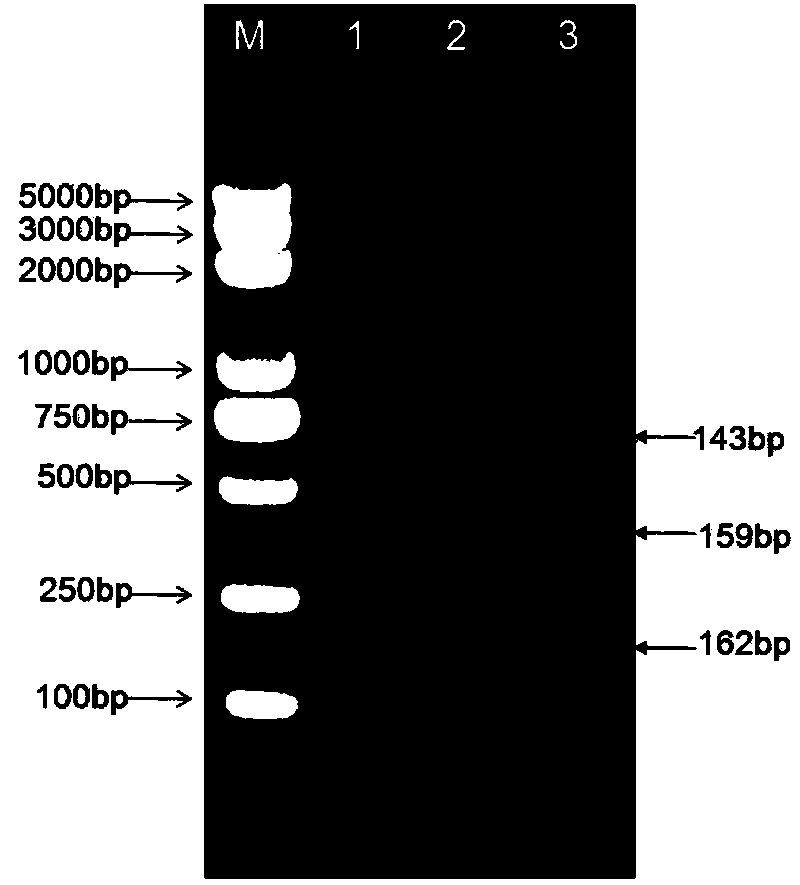
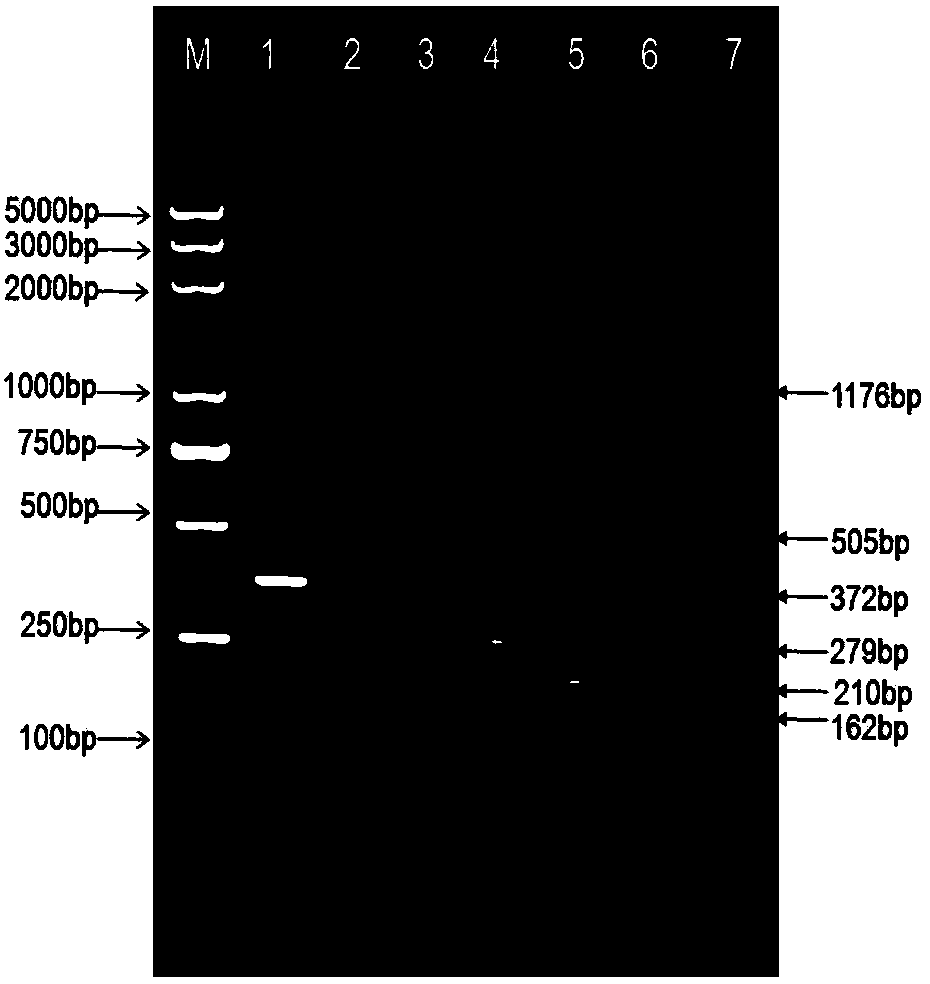
Shigella flora/serotype multiplex-PCR (polymerase chain reaction) detection primer set and kit
A Shigella, Shigella genus technology, applied in the field of microbial detection, can solve the problems of Shigella missed detection, expensive biochips, poor stability, etc., to save time, reduce labor costs and time costs, The effect of avoiding missed detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0077] Example 1 Design and synthesis of primers for Freund's type 6 O antigen encoding gene wzx
[0078] Search for 10 full-length sequences of Freund's type 6 O antigen-encoding gene wzx (Freund's type 6) in Genbank, import 10 full-length sequences of wzx (Fundi's type 6) gene into Mega4 software, and use the alignment function for alignment The conservative sequence of wzx (Freund 6) gene was analyzed, and the conservative sequence segment was obtained. The base sequence is shown in SEQ ID No. 14 in the sequence table.
[0079] Input the wzx (Frankenstein type 6) gene sequence into primer premier6 software, and automatically analyze and obtain multiple primer design schemes. On this basis, manually adjust the primer position and sequence length, set the Tm value to 55±5°C, and the GC content of 35%-65%. As far as possible, no hairpin structure and primer dimers are produced, and no error is caused. Perform the Blast analysis on the above-mentioned manually adjusted primer desig...
Embodiment 2
[0083] Example 2 Screening test of Shigella detection primers
[0084] Screening of wzx (Freund 6) gene primer options in Table 2 mainly evaluates the specificity and sensitivity of the primers.
[0085] Specificity evaluation: Extract Escherichia coli ATCC25922, Enterobacter sakazakii, Staphylococcus aureus, Salmonella, Vibrio parahaemolyticus, Listeria monocytogenes, Vibrio cholerae, Bacillus cereus, jejunum Total DNA of Campylobacter, Campylobacter coli, Enterobacter sakazakii, Yersinia enterocolitica, Aeromonas hydrophila, etc. Take 10μL of extract for each bacterium and mix it into a comprehensive DNA template as a specific As a template for verification, the DNA content was determined to be 183.37ng / μL using Nanodrop.
[0086] Dilute the primers into 100μM stock solution with TE (pH8.0) solution, and then prepare the corresponding solution. Configure the reaction system as follows: Taq DNA polymerase 0.4μL (5U / μL), 5×PCR buffer (Tris·HCl100mM (PH8.3), KCl250mM, tween-200.2%) ...
Embodiment 3
[0098] Example 3 Establishment of multiple PCR detection kit for identification of Shigella flora and serotype
[0099] The kit consists of 2× reaction system buffer, DNA polymerase, 10× primer mixture, positive control, and deionized water. The specific components are as follows: 2×PCR Buffer (Tris·HCl40mM (PH8.3), KCl100mM , Tween-200.08%, 0.0006ng / μL pET28a, 1mm dNTP, 8mm MgCl 2 ); 25×DNA polymerase (2U / μL); 10× primer mixture (each primer concentration including IAC primers is 2um), positive control (Freund 2a, Freund 6 and Dysentery I Mixed template with Shigella Sonnei, 10 each 6 CFU / mL).
[0100] The reaction system tested by the kit is 25μL, and the configuration is as follows: 2×PCR Buffer 12.5μL; 25×DNA polymerase 1μL; 10×primer mixture 2.5μL; template 2μL, deionized water 7μL.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com