Puccinia striiformis quantitative detection kit and application thereof
A technology for quantitative detection of wheat stripe rust, applied in the direction of microbial measurement/inspection, microbial-based methods, microorganisms, etc., can solve the problems that have not yet been reported on the early quantitative monitoring method of wheat stripe rust, and achieve reliable detection means, High amplification efficiency and low detection limit
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] Embodiment 1, employing fungal conserved gene primers to analyze wheat rust
[0054] With reference to the nucleotide sequence of Bt2a / Bt2b reported in "GlassandDonaldson, 1995.":
[0055] Bt2a: 5'-GGTAACCAAATCGGTGCTGCTTTC-3' (Seq ID No. 1),
[0056] Bt2b: 5'-ACCCTCAGTGTAGTGACCCTTGGC-3' (Seq ID No. 2).
[0057] The PCR analysis of gDNA of wheat stripe rust, leaf rust fungus and stem rust fungus was carried out on the PTC2220 PCR instrument of MJResearch, Inc.
[0058] The amplification system is 25 μL, including 10×PCRBuffer (Mg2+Plus) 2 μL, dNTPMixture (2.5 mmol / L each) 0.3 μL, Bt2a / Bt2b (10 μmol / L) 1 μL, template DNA (20ng / μL) 1.0 μL, TaKaRaTaq (5U / μL) 0.3 μL, ddH2O 16.9 μL.
[0059] The PCR reaction conditions were: 94°C for 2 min, 1 cycle; 94°C for 15 s, 55°C for 30 s, 72°C for 90 s, 30 cycles; 72°C for 10 min, 1 cycle.
[0060] The amplified products were separated by agarose gel with a mass fraction of 1.5 and 0.5×TBE electrophoresis buffer, and the electrop...
Embodiment 2
[0062] Embodiment 2, the preparation of kit of the present invention
[0063] 1. Acquisition of primers
[0064] According to the stripe rust specific sequence SeqID No.3 obtained in Example 1, a series of primers for fluorescent quantitative PCR detection were designed using the software Primer5.0.
[0065] After primer screening and verification, the specific primers in the kit of the present invention are finally obtained, and the nucleotide sequences of the forward and reverse primers are as follows:
[0066] osgQ607 / osgQ608:
[0067] GGATGCCTCGAAGGGTTATACC (Seq ID No. 5) / TGCTAGATACAATGGCACATCTGA (Seq ID No. 6).
[0068] 2. Kit Assembly
[0069] The primers, 2×SGPCRMasterMix, 2×SGGreenqPCRMix (withROX) and double-distilled water are combined according to a certain dosage ratio to assemble the kit described in the present invention.
Embodiment 3
[0070] Embodiment 3, kit of the present invention detects wheat sample
[0071] step:
[0072] A. Obtain standard curve
[0073] 1. Preparation of DNA standard: Extract the genomic DNA (plasmid DNA) of the stripe rust CY33 strain according to the operating instructions of the plasmid DNA extraction kit, and use the plasmid DNA as a template to add primers osgQ607 / osgQ608 and common PCR amplification reagents (as in Table 1) PCR amplification; the reaction system of the PCR amplification is as shown in Table 1; the reaction program is as shown in Table 2; the amplified stripe rust PCR product is the DNA standard.
[0074] Table 1
[0075]
[0076]
[0077] Table 2
[0078]
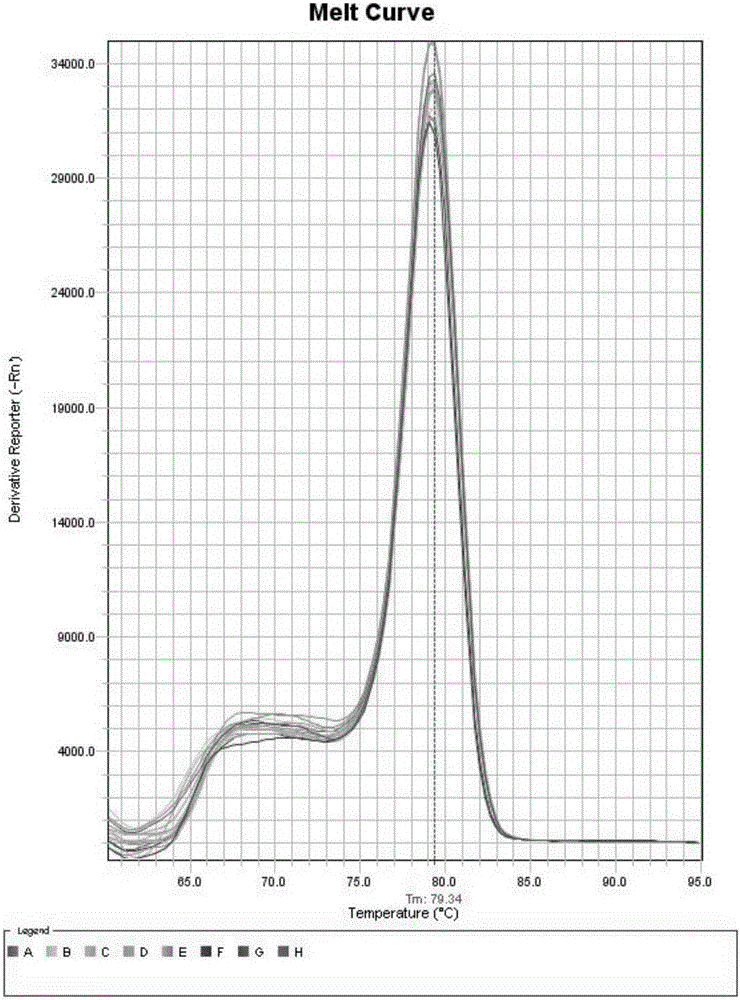
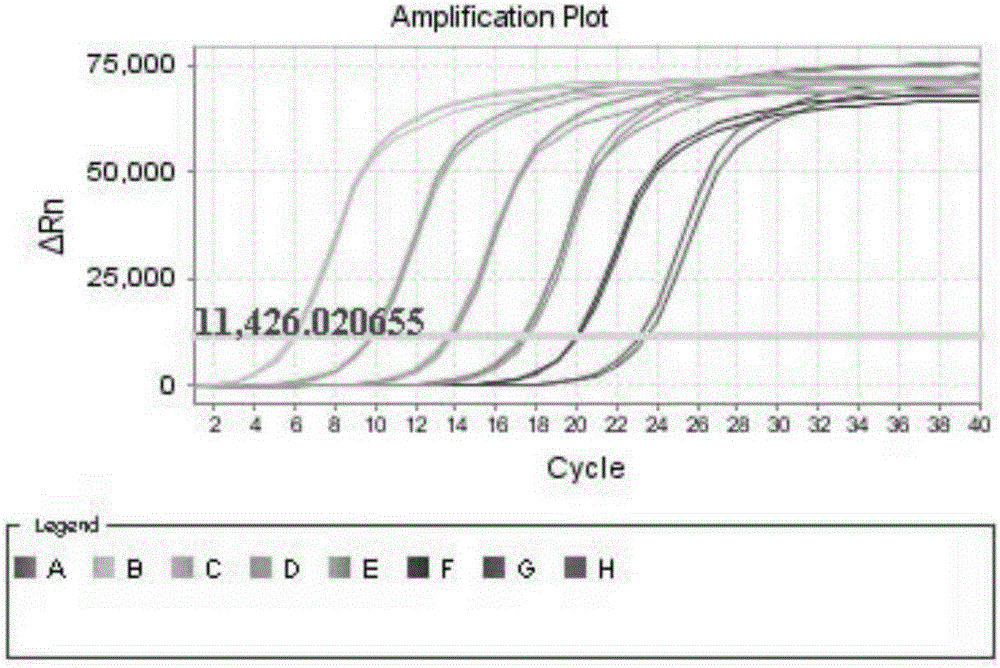
[0079] 2. Use a spectrophotometer to measure the concentration of the above PCR product to be 498ng / μl, carry out gradient dilution according to a tenth ratio to form a series of DNA standard dilutions, and calculate the copies of the serial dilutions according to the gradient concentration an...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com