A mutant gene of the gshf gene of Streptococcus agalactiae and its application
A technique for mutating genes and Streptococcus lactis, which is applied in the field of bioengineering to achieve the effects of reducing Km value, improving catalytic efficiency and enhancing affinity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
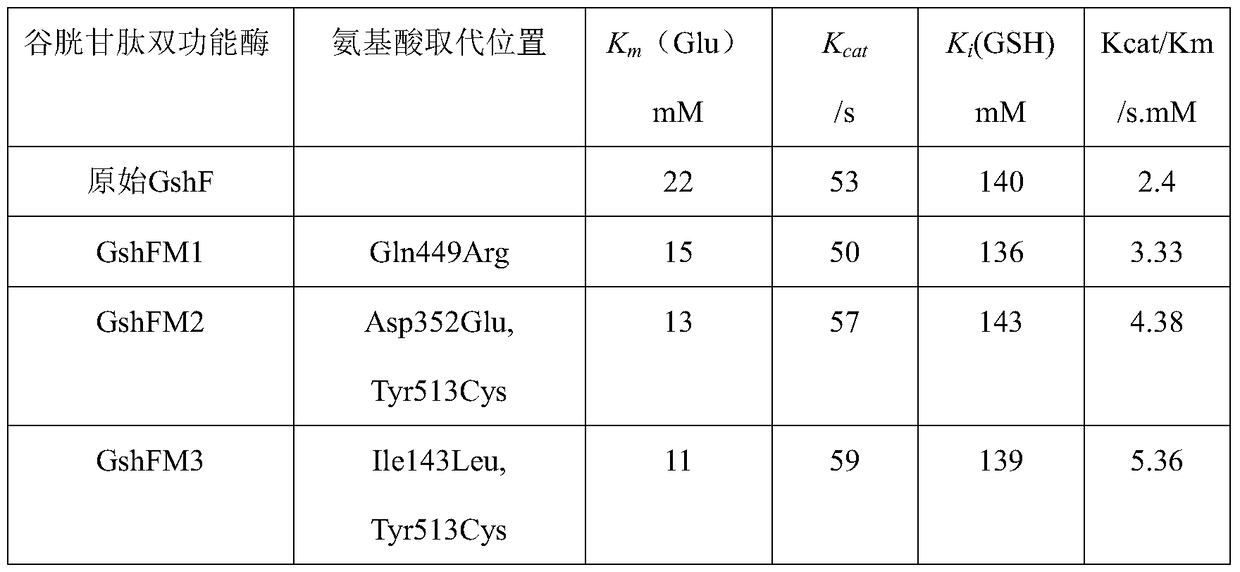
[0027] In this example, an error-prone PCR method was used to construct a mutant library of the glutathione bifunctional enzyme gene, and to screen glutathione bifunctional enzyme mutants with high enzymatic activity.
[0028] 1. Construction of recombinant plasmid pUC19-gshF.
[0029] Template: Streptococcus agalactiae S. agalactiaeATCC-BAA611 genomic DNA;
[0030] Vector: PUC19 was digested with HindⅢ / PstⅠ, purified and recovered to obtain the linearized plasmid PUC19. The PUC19 is purchased from Takara Company with a catalog number of 3219.
[0031] Primers:
[0032] gshF upstream primer gshF-1:CCC AAGCTT ATGATTATCGATCAACTGTTAC;
[0033] gshF downstream primer gshF-2: TGCA CTGCAG TTATAATTCTGGGAACAGTTTA;
[0034] PCR amplification conditions: 94°C for 5min, 94°C for 30s, 65°C for 30s, 72°C for 2.5min, a total of 30 cycles.
[0035] The amplified PCR product was digested with HindⅢ / PstⅠ, and then ligated with the above-mentioned linearized plasmid PUC19 after gel cutt...
Embodiment 2
[0067] In this example, the glutathione synthase gene mutant obtained in Example 1 was used to prepare Pichia pastoris engineering bacteria.
[0068] 1. The mutant genes gshFM1, gshFM2, gshFM3 and gshF were obtained by PCR reaction.
[0069] Template: the recombinant plasmids PUC19-gshFM1, PUC19-gshFM2, PUC19-gshFM3 obtained above and the genome of Streptococcus agalactiae ATCC-BAA611
[0070] Primers:
[0071] Upstream: 5'-CAATTGAACAACTATTTCGAAACGATGATTATCGATCAACTGTTACAAAGAAG-3';
[0072] Downstream: 5'-GACGGCCGGCTGGGCCACGTGAATTTATAATTCTGGGAACAGTTTAGCCAA-3';
[0073] PCR amplification reaction: 1 μL of template, 2 μL of 10×PCR buffer, 2 μL of upstream and downstream primers, 5 μL of dNTP, 0.5 μL of TaqDNA polymerase, the total reaction volume is 20 μL, PCR cycle program: 94°C for 5min, 94°C for 30s, 65°C for 30s , 72°C for 2.5min, a total of 30 cycles. The PCR products were recovered by agarose gel electrophoresis and then digested with Cla Ⅰ and Not Ⅰ to obtain glutathio...
Embodiment 3
[0079] In this example, the Pichia pastoris engineered bacteria obtained in Example 2 were used to synthesize glutathione.
[0080] Inoculate single colonies of GS115 / gshFM1, GS115 / gshFM2, GS115 / gshFM3, GS115 / gshF and GS115 grown on YPD medium solid plates for three days at 30°C into 30mL of YPD medium, at 30°C, 220rpm Cultivate for 20 hours; then put it into 320mL of YPD medium, put it into 6L (10L fermenter) fermentation medium BMGY after 8h, add defoamer, control the temperature at 30°C, add glycerol during the process, and control the pH at the same time In 6.0. Start to add cysteine after 30 hours of fermentation, keep the concentration of cysteine in the fermentation broth at 0.1-0.2mmol / L, and continue to ferment until 70 hours to stop fermentation. The YPD medium components are Glucose 20g / L, Tryptone 20g / L, Yeast Extract 10g / L. The composition of the BMGY medium is: yeast 10g / L, peptone 20g / L, yeast nitrogen source 1.34g / L, 0.1M pH6.0 phosphate buffer 10mL / L, gl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com