Alkaline beta-mannase, encoding genes thereof and application of encoding genes
A mannanase, alkaline technology, applied in the field of genetic engineering, can solve the problems of low enzyme activity, low expression, poor stability, etc., and achieve the effect of high activity, high activity and strong stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
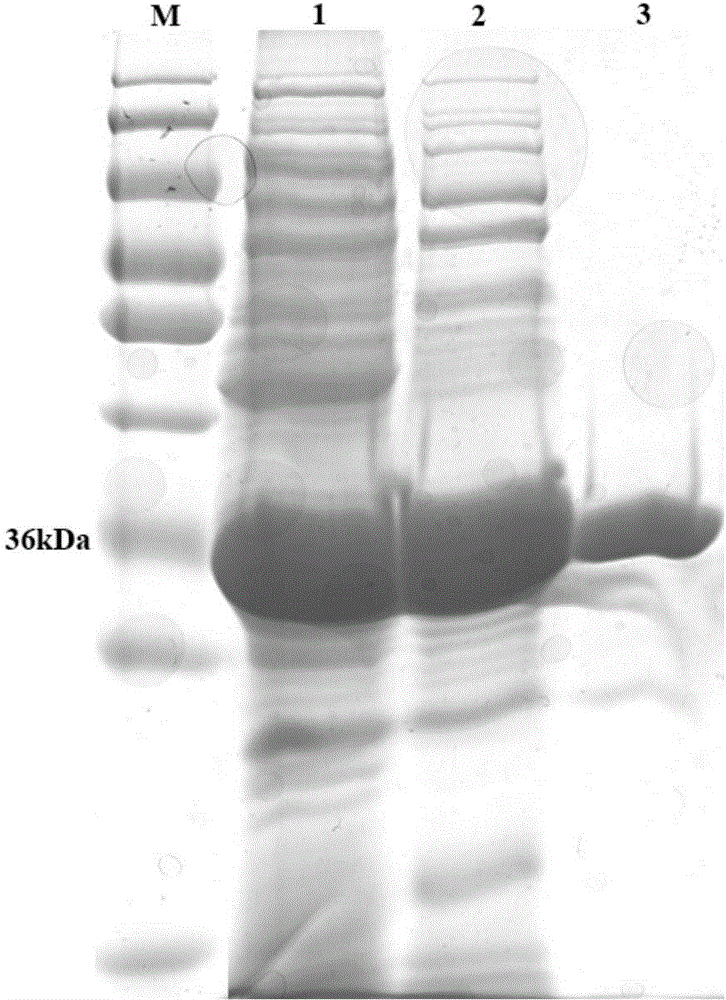
[0048] The method for preparing alkaline β-mannanase provided by the invention comprises: cultivating the recombinant strain provided by the invention, inducing the expression of the gene encoding alkaline β-mannanase; separating and purifying the expressed alkaline β-mannanase Glycanase. The culture conditions are conventional culture conditions, such as using LB medium (the solvent is water, the solute and its final concentration are respectively: Tryptone10g / L, yeast extract 5g / L, NaCl10g / L), at 35-37°C Grow to OD 600 is 0.6. Since the recombinant strain provided by the invention contains the gene encoding the alkaline β-mannanase, it can efficiently express the alkaline β-mannanase. After culturing, the high-purity alkaline β-mannanase can be obtained through separation and purification. Methods known to those skilled in the art can be used for separation and purification (for example, adding isopropyl-β-d-thiogalactopyranoside (IPTG) to the culture medium to a final co...
Embodiment 1
[0055] Acquisition of alkaline β-mannanase and its coding gene
[0056] (1) Cloning of the gene encoding alkaline β-mannanase (ManA)
[0057] Take the Bacillus clausii S10 isolated from the alkaline hot spring sample in Inner Mongolia, use the genome extraction kit to extract the total DNA of the Bacillus clausii S10, and measure the purity of the DNA with a UV spectrophotometer. The result is: A260 / A280 = 1.88, A260 / A230 = 2.13. 10 μl of the total DNA solution (about 50 μg DNA) was taken, partially digested with restriction endonuclease Sau3AI, and a 2-8 kb DNA fragment was recovered by agarose gel electrophoresis. Then carry out ligation reaction, 4°C ligation reaction for 16 hours, the ligation system is as follows (20 μl):
[0058]
[0059]
[0060] Transform competent Escherichia coli DH5α with the product of the ligation reaction, and then apply it to a solid at pH 8.0 containing 60 μg / ml ampicillin (Amp), 20 μg / ml IPTG, 40 μg / ml galactoside (X-gal) and 0.5% konj...
Embodiment 2
[0076] Detection of Enzymatic Properties of Alkaline β-Mannanase (ManA)
[0077] (1) Standard enzyme activity assay method
[0078] Take 10 μl of the ManA enzyme solution obtained in Example 1 (diluted to 1.5 μg / ml) and 190 μL of glycine-NaOH buffer solution with a pH value of 9.5 containing 0.4% locust bean gum, mix well, react at 75°C for 10 minutes, and add 200 μL Dinitrosalicylic acid solution (DNS) terminated the reaction (edited by Zhang Longxiang et al., "Biochemical Experimental Methods and Techniques", Higher Education Press, 1996), and then measured the absorbance at 540nm after reacting in a boiling water bath for 5 minutes.
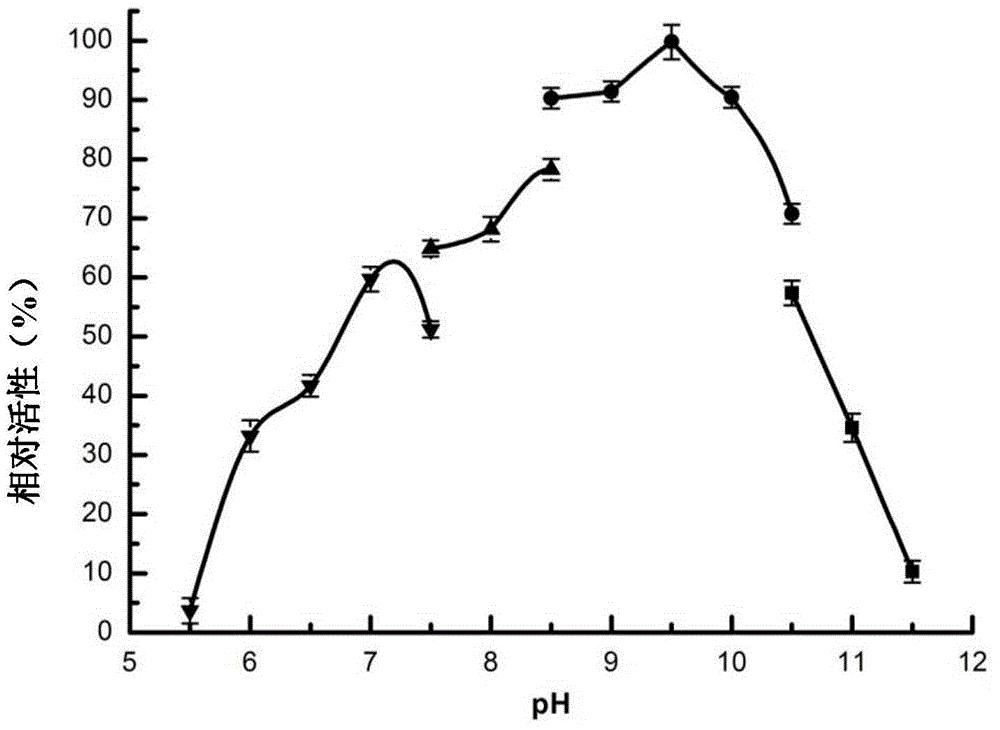
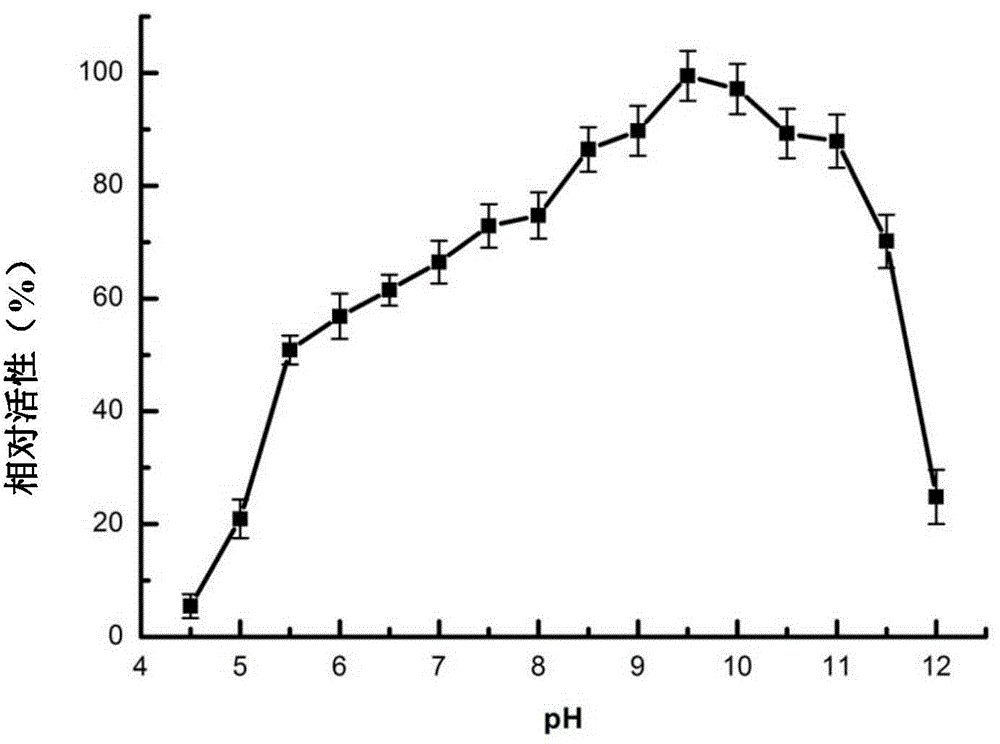
[0079] (2) Determination of the optimum pH value and pH stability of ManA
[0080] At 75°C, the ManA enzyme solution was subjected to enzymatic reactions in buffer solutions with different pH values (pH5.5-11.5) to determine its optimum pH value. The remaining conditions were the same as (1), and the buffer solution used was pH5.5 -7.5Na ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Specific vitality | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com