Heatproof mutation lipase with high catalytic activity as well as preparation method and application of heatproof mutation lipase
A high catalytic activity, lipase technology, applied in the field of enzyme engineering, can solve the problems of not paying attention to the protein unfolding mode, not screening out the key subregions of unfolding, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
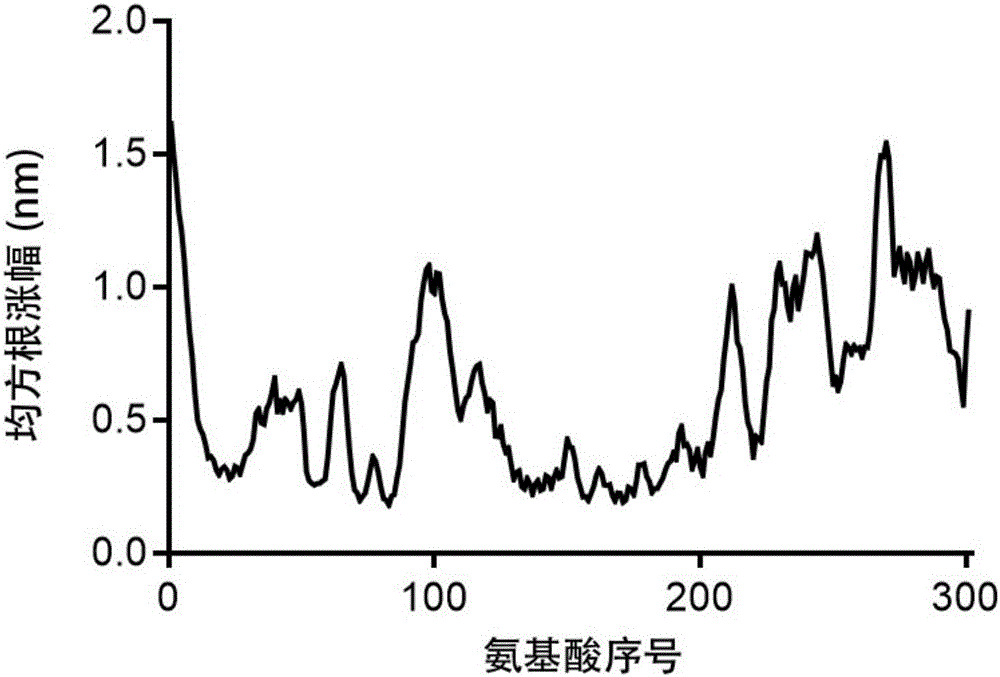
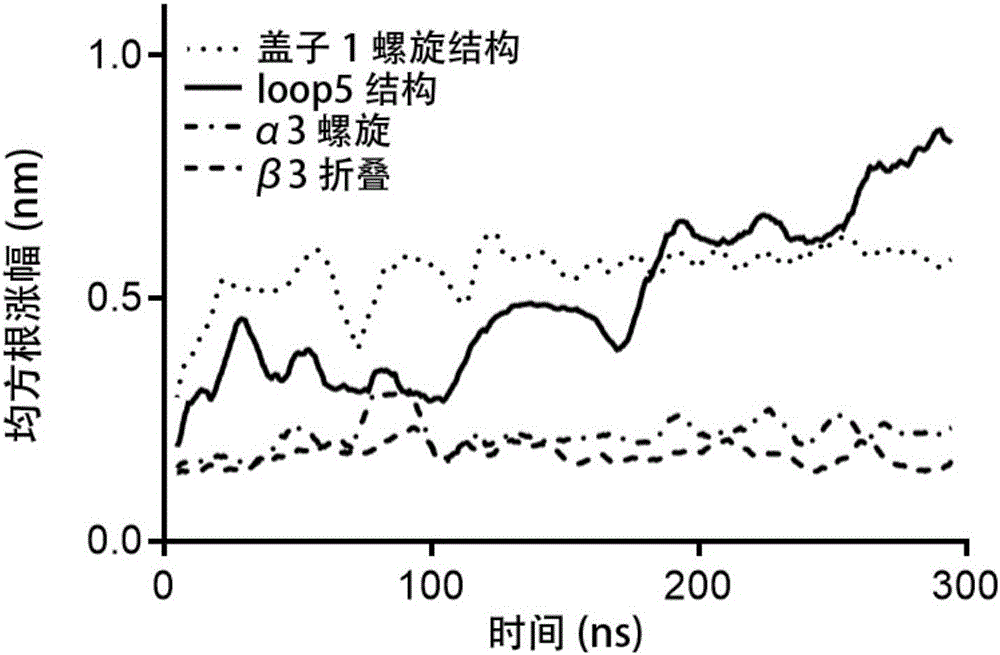
[0050] Example 1 Establishment of unfolding model of Yarrowia lipolytica lipase 2 and screening of disulfide bonds.
[0051] (1) Search and download the crystal structure of Lip2 (PDB ID: 3O0D) from the RCSB PDB crystal database with an accuracy of The crystal is a heptamer, and each polymer has different degrees of missing atoms. Therefore, the complete A-chain of the skeleton atoms is selected as the initial model, and the missing side chain amino acids of the A-chain are repaired using the Swiss-pdb Viewer software, and Disulfide is used to By Design software, the Cys120-Cys123 disulfide bond in the A chain is reconstructed, and all the crystal water of the A chain is retained in the simulation. Except for the HIS289 in the center of the triplet, the HID protonation state is used, and the rest of the histidines are set to Gromacs by default. The protonation state of pH=7.00, the pretreatment model is obtained;
[0052] (2) Use the GROMACS 5.04 software to carry out three-...
Embodiment 2
[0083] Construction of embodiment 2 mutant lipase expression plasmid
[0084] Using the Yarrowia lipolytica lipase 2 sequence (Genbank ID: AJ012632.1) as the target fragment, with EcoRI and NotI as restriction sites, the whole gene was synthesized and pPICZαA-Lip2 was constructed. The second serial inverse PCR method introduces disulfide bond mutations.
[0085] 105C-F and 105-R, 112C-F and 112-R, 118C-F and 118C-R, 122C-F and 122C-R, 125C-F and 125C-R in Table 2 were used as primers to obtain S105- Primers for the first amplification of 124, S112-137, S118-177, S122-196 and S125-193.
[0086] The PCR amplification conditions are: 94°C for 2min; 94°C for 10s, 66°C for 30s, 68°C for 5min, 10 cycles. The reaction system is shown in Table 3 below.
[0087] The amplified product was digested with DnpI enzyme to digest the template, and after agarose gel electrophoresis to detect the size of the mutant band, it was ligated and circularized overnight with T4 ligase, and then the ...
Embodiment 3
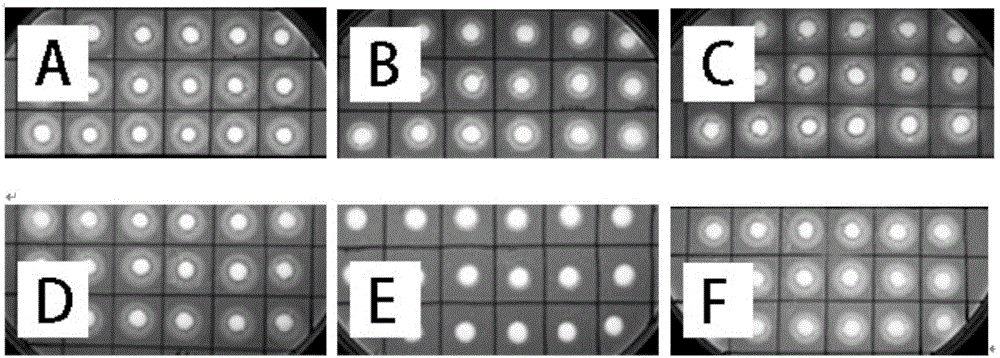
[0095] Example 3: Electrotransformation of Pichia pastoris with linearized plasmid, screening of transformants and screening of enzyme production
[0096] After the positive transformants with correct sequencing were expanded overnight in LLB+Zeocin (Zeocin concentration: 25 μg / ml) liquid medium, the plasmid was extracted, linearized with PmeI, purified and recovered, and a total of 5 μg of the plasmid linearized product was mixed with Mixed electric shock transformation of competent X33 Pichia pastoris. Competent Pichia pastoris was prepared according to the operation manual of Invitrogen Company. The electroporation program was set according to the parameters recommended by Bio-Rad.
[0097] Add 1 mL of 1mol / L sorbitol solution immediately after electroporation, incubate and recover the bacterial solution at 30°C for 1 hour, and spread it evenly on the YPDS+Zeocin (Zeocin concentration is 200 μg / ml) resistance plate for screening; after culturing for 3 days, put The grown ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com