Degenerate primers for identifying hybrid isoprenoid-producing bacteria and application thereof
A technology of isoprenoid and degenerate primers, applied in the biological field, can solve problems such as blindness, time-consuming, and tediousness, and achieve the effects of strong purpose, avoiding tediousness, and improving work efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
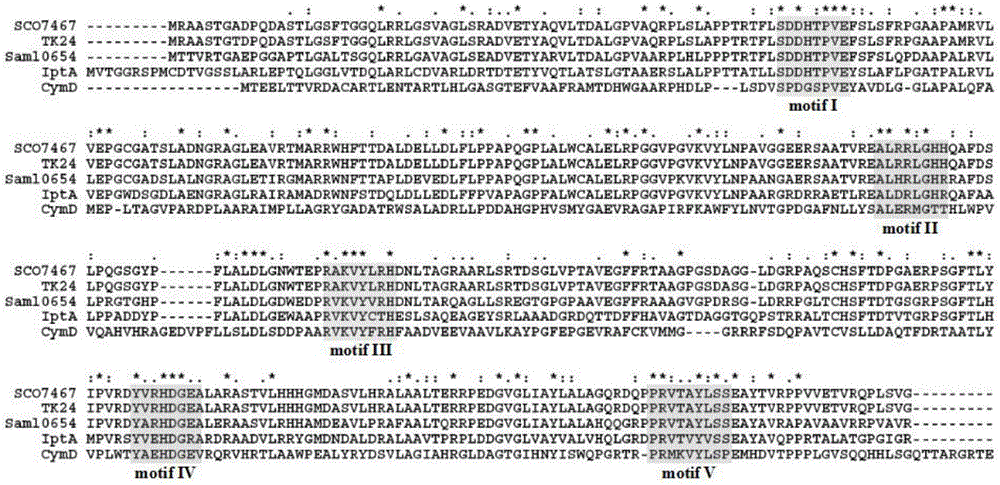
[0039] Example 1 Design of degenerate primer pairs
[0040]Due to the phenomenon of degeneracy of amino acid codons, that is, the amino acid sequence of the final protein expressed by different DNA sequences may be the same, so comparing the published protein sequences of prenyltransferases, primer pair 1 was based on the conservative Design degenerate primers for region motif I and motif II. For primer pair 2, the upstream and downstream primers were designed based on the conserved regions of prenyltransferase, motif I and motif V, and for primer pair 3, the upstream and downstream primers were designed based on the conserved regions of prenyltransferase, motif I and motif IV.
[0041] The protein sequences of isopentenyltransferases from different microorganisms were collected from NCBI, Streptomycessp.SN-5936-dimethylallyltryptophan synthase (GenBank number: BAJ07990.1), Streptomyces coelicolor A3(2) SCO7467 (GenBank number: CAC36059. 1), Streptomyces lividans TK24 (GenBan...
Embodiment 2
[0049] Example 2 Extraction of microbial DNA
[0050] Five heterozygous isoprenoid-producing strains were taken: Streptomyces sp.neau-D50, Streptomyces scoelicolor A3(2), Streptomyces lividans TK24, Streptomyces ambofaciens ATCC23877, Salinispora arenicola CNS-205, and their respective genomes were extracted.
[0051] The extraction method of microbial DNA is as follows: (1) add 5 ml of SET buffer solution to the centrifuge tube containing microbial cells, pipette and mix to resuspend the bacteria, then add 100 μl lysozyme solution (50 mg / ml) to a final concentration of 1mg / ml, incubate in a 37°C water bath for 30-60min until the bacterial suspension is viscous and transparent; (2) Slowly add 140μl proteinase K solution (20mg / ml) to the bacterial suspension to a final concentration of 0.5mg / ml, Mix by inverting, then add 600 μl of 10% SDS solution to a final concentration of 1%, mix by inverting, then incubate in a water bath at 55°C for 2 hours, and mix by inverting slowly ev...
Embodiment 3
[0052] Example 3 Verification of Primer Validity and Accuracy
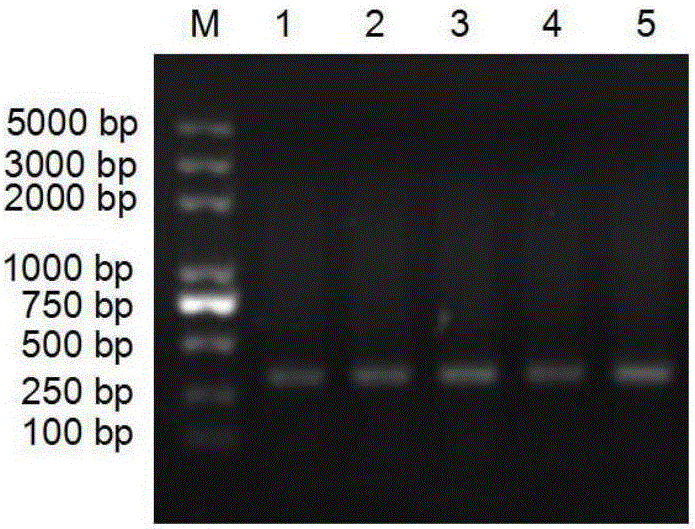
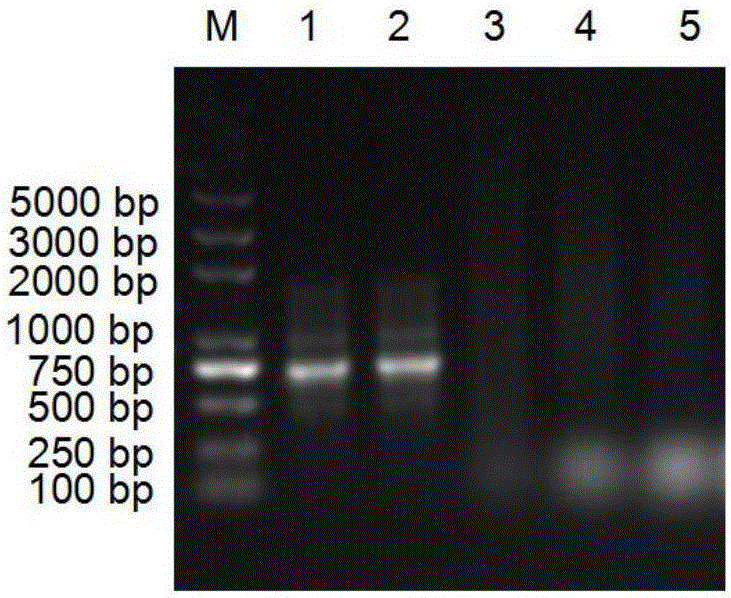
[0053] The three sets of degenerate primers designed in Example 1 were used to perform PCR on each genome, and then detected by electrophoresis. The PCR product was recovered, and the PCR product was sent to Bao Biological Engineering (Dalian) Co., Ltd. for sequencing. PCR reaction system: 10 μl of 10× amplification buffer, 200 μmol / L of each dNTP mixture, 10-100 pmol of each primer, 0.1-2 μg of template DNA, 1.5 U of Taq DNA polymerase, Mg 2+ 1.5mol / L, add deionized water to 100μL. The amplification reaction conditions are: 94°C for 10 min; 94°C for 1 min; 53°C for 30 s; 72°C for 1 min.
[0054] As a result, it was found that the PCR product of primer pair 1 had a single band at 357bp (such as figure 2 shown), indicating that the primers can indeed be used to amplify the isopentenyl transferase gene from the heterozygous isoprenoid compound-producing bacteria. The sequencing results of the PCR products were ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com