Polypeptide having pNPPC hydrolase activity and coding gene, preparation method and application thereof
A technology for hydrolyzing activity and residues, which is applied in the field of polypeptides with pNPPC hydrolase activity, and can solve the problems of large interference, low activity, and easy false positives of the blank control
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0142] Example 1: Fermentation of wild pNPPC hydrolase capable of hydrolyzing pNPPC to produce pNP
[0143] A ring of activated Bacillus licheniformis CGMCC No.7878 grown on PDA or LB agar medium was inoculated into a seed shaker flask, and cultured at 28° C. and 180 rpm for 24 hours. The seed medium is LB liquid medium, which consists of 1% tryptone, 0.5% yeast extract, 1% sodium chloride, and pH 7.0.
[0144] After the liquid seeds are cultured, they are inoculated into 30 ml fermentation medium with an inoculation amount of 1%, and cultured at 28° C. and 180 rpm for 24 hours. The composition of the fermentation medium is: 0.5% sucrose, 0.5% tryptone, 1% yeast extract powder, 0.5% beef extract, 0.5% corn steep liquor, K 2 HPO 4 0.05%, MgSO 4 0.05%, CaCl 2 0.05%, ZnSO 4 ·7H 2 O0.05%, manganese sulfate 0.1%, pH 7.0.
[0145] After the cultivation, the culture solution was centrifuged at 1200 rpm for 10 minutes, and about 800 ml of centrifuged supernatant was collected, ...
Embodiment 2
[0146] Embodiment 2: Separation and purification of wild pNPPC hydrolase
[0147] About 800ml of the centrifuged supernatant in Example 1 was microfiltered with a 0.22 μm microfiltration membrane, then concentrated by ultrafiltration with a 10KDa ultrafiltration membrane, and replaced with 20mM Tris-HCl pH8.7 buffer. Collect ultrafiltration retentate about 100ml.
[0148] Use a 5ml DEAE column (Hitrap TM DEAE FF, GE Healthcare) carried out anion exchange chromatography separation on the ultrafiltrate of above-mentioned about 100ml volume. The elution buffer was 20 mM Tris-HCl pH 8.7 buffer containing 1 M NaCl. The conditions of ion exchange chromatography are as follows:
[0149] Buffer A: 20mM Tris-HCl, pH8.7;
[0150] Buffer B: 20mM Tris-HCl, 1M NaCl, pH8.7;
[0151] Flow rate: 2ml / min;
[0152] Elution method: Gradient elution 0-100% B, 120min;
[0153] Collection: 2ml / tube.
[0154] The collected eluate was tested for pNPPC activity, and the collected solution wit...
Embodiment 3
[0163] Example 3: Mass spectrometry identification of wild pNPPC hydrolase protein
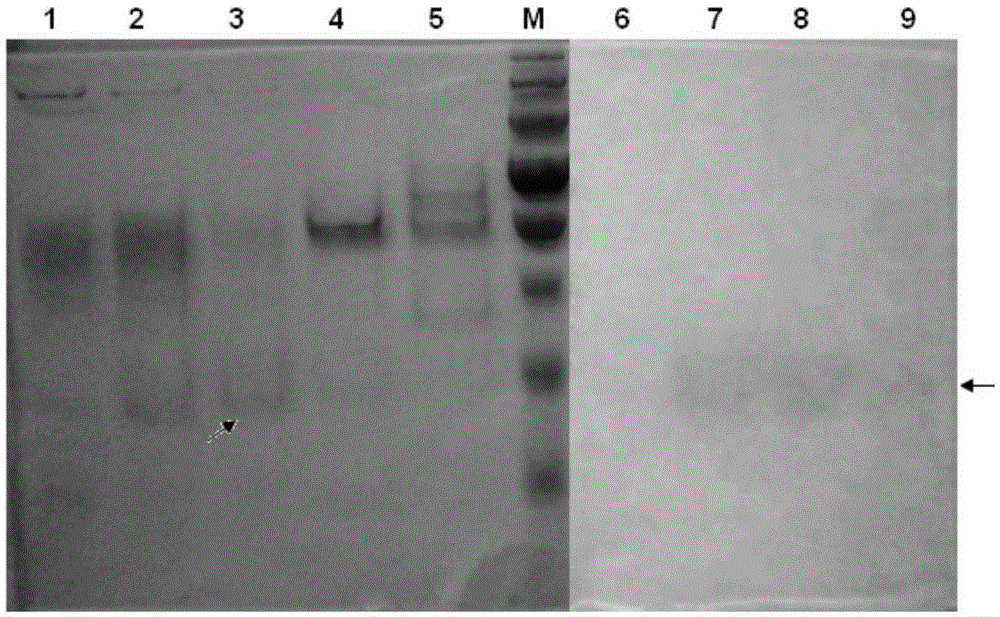
[0164] Figure carefully cut out with a clean blade figure 1 The protein band in lane 3 pointed to by the oblique arrow was then identified by LC-MS / MS mass spectrometry. After data analysis, it was found that the protein with the highest score in this band was a protein in the Bacillus licheniformis genome that was speculated to be 2',3'-cyclic nucleotide 2'-phosphodiesterase, and its GENE BANK number was WP_016886260. The polynucleotide sequence of the protein is shown in SEQ ID NO:11, and the amino acid sequence is shown in SEQ ID NO:12. The predicted protein has a length of 1445 amino acids and a molecular weight of 158.2 KDa, which is much higher than the molecular size of the wild pNPPC hydrolase prepared in Example 2.
[0165] At the same time, the mass spectrometric identification results of LC-MS / MS also gave several very high-scoring peptides, and their amino acid sequence informati...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Apparent molecular weight | aaaaa | aaaaa |
| Molecular weight | aaaaa | aaaaa |
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com