Biosensor for detecting kanamycin and preparation method thereof
A biosensor, kanamycin technology, applied in the field of biosensors, can solve the problems of high cost, low specificity and sensitivity, and achieve the effects of simple electrode, improved sensitivity and stable performance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
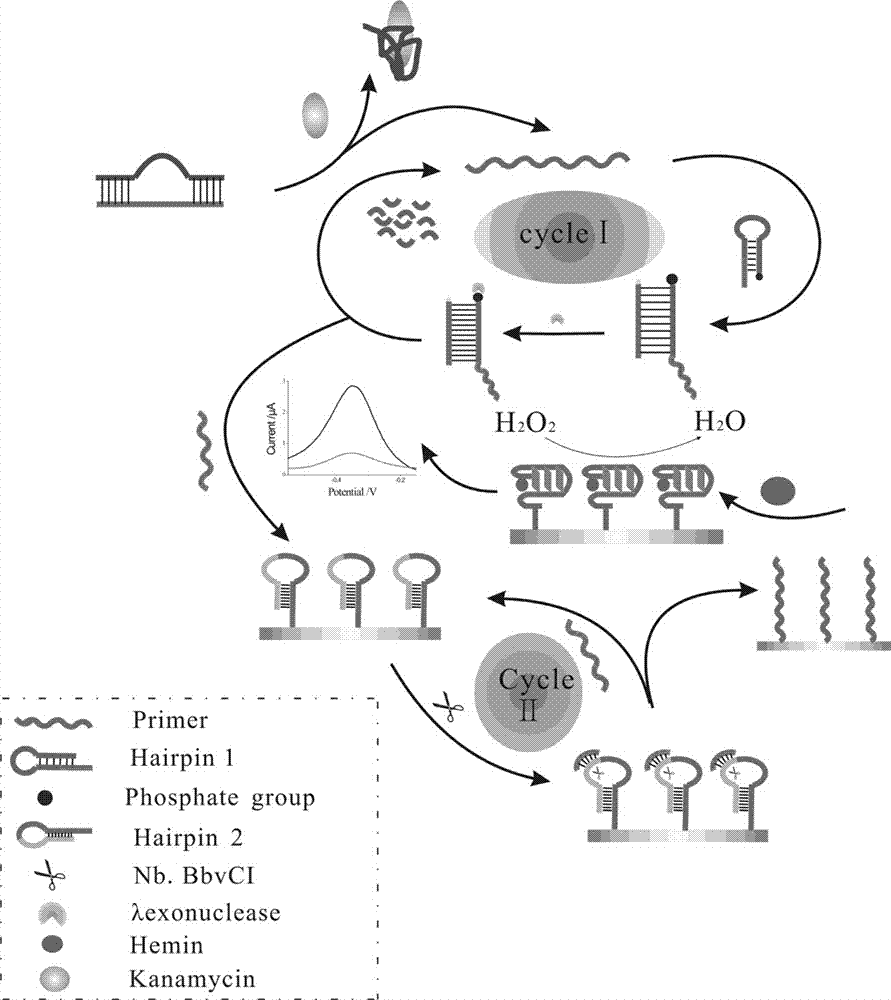
[0045] The preparation method of described biosensor comprises the following steps:
[0046] (1) Pretreatment of the electrodes;
[0047](2) Modification of HAP2 to the electrode surface;
[0048] (3) Modification of the homogeneous reaction product layer onto the electrode surface.
[0049] In the preparation method, the operation steps of modifying HAP2 onto the electrode surface are preferably as follows: 10 μL of HAP2 is added dropwise to the pretreated electrode surface, and incubated at 37° C. for 2 hours.
[0050] In the preparation method, the preferred steps of modifying the homogeneous reaction product onto the electrode surface are as follows:
[0051] (1) Add sterilized water, 10× buffer buffer, HAP1, Primer, Helper, Lambda exonuclease, and the target to be tested into a centrifuge tube, shake for 30 seconds, and incubate in a 37°C incubator for 2 hours;
[0052] (2) Place the incubated mixed solution in an incubator at 75°C for 5 minutes to inactivate Lambda ex...
Embodiment 1
[0059] The main steps of the electrode modification process are as follows:
[0060] a. The gold electrode is first polished in 0.3 and 0.05 µm alumina slurry until it becomes a mirror surface, and then rinsed repeatedly with PBS and secondary water;
[0061] b. Add 10 μL of HAP2 (10 μM) dropwise to the electrode surface and incubate at 37°C for 2h. Fix the sulfhydryl chains to the electrode surface through Au-S bonds;
[0062] So far, the modification process of the electrode has come to an end. The following describes the reaction in the homogeneous solution and the main steps in the homogeneous reaction:
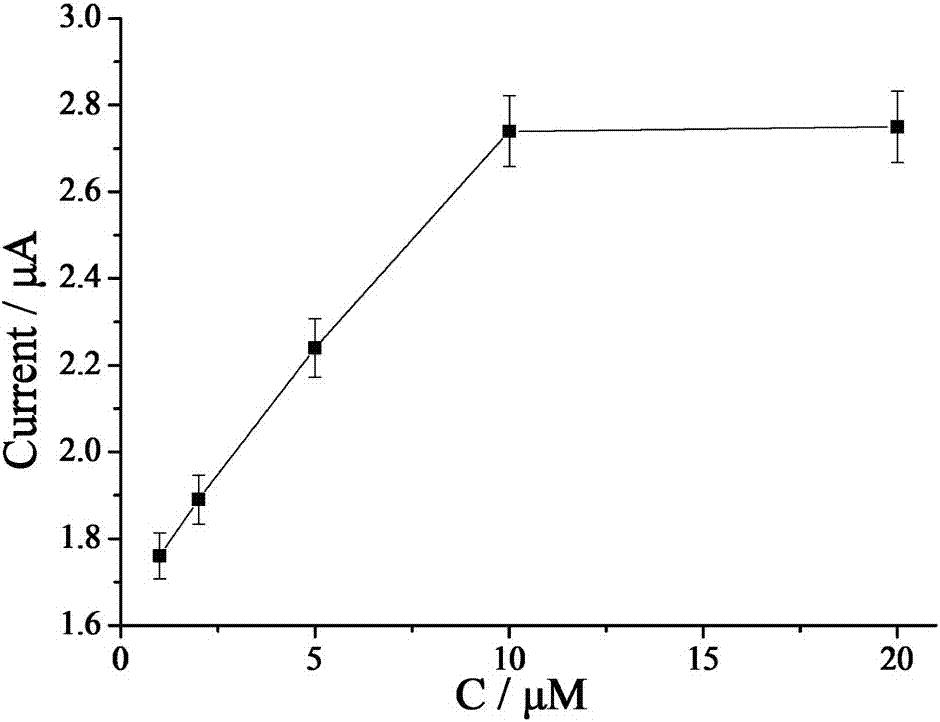
[0063] a. Mix 3 μL sterilized water, 5 μL 10× buffer buffer, 4 μL different concentrations of HAP1 (5 μM, 10 μM, 25 μM, 50 μM, 75 μM, 100 μM), 2 μL Primer (10 μM), lambda exonucleic acid Enzyme (1 μL), 2 μL Helper (50 μM), and 2 μL target substance to be tested were added to a centrifuge tube, shaken for 30 seconds, and incubated in a 37°C incubator for 2 hours;
[006...
Embodiment 2
[0076] The main steps of the electrode modification process are as follows:
[0077] a. The gold electrode is first polished in 0.3 and 0.05 µm alumina slurry until it becomes a mirror surface, and then rinsed repeatedly with PBS and secondary water;
[0078] b. Add 10 μL of HAP2 (10 μM) dropwise to the surface of the electrode and incubate at 37°C for 2 hours. Fix the sulfhydryl chains to the electrode surface through Au-S bonds;
[0079] So far, the modification process of the electrode has come to an end. The following describes the reaction in the homogeneous solution and the main steps in the homogeneous reaction:
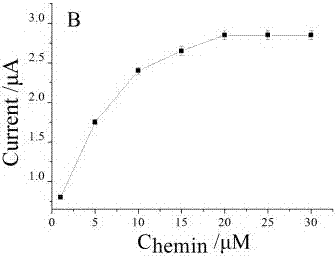
[0080] a. Mix 3 μL sterilized water, 5 μL 10× buffer buffer, 4 μL different concentrations of HAP1 (50 μM), 2 μL Primer (10 μM), lambda exonuclease (1 μL), 2 μL Helper (100 μM), and Add 2 μL of the target substance to be tested into a centrifuge tube, shake for 30 seconds, and incubate in a 37°C incubator for 2 hours. b. Incubate the incubated mixed solutio...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com