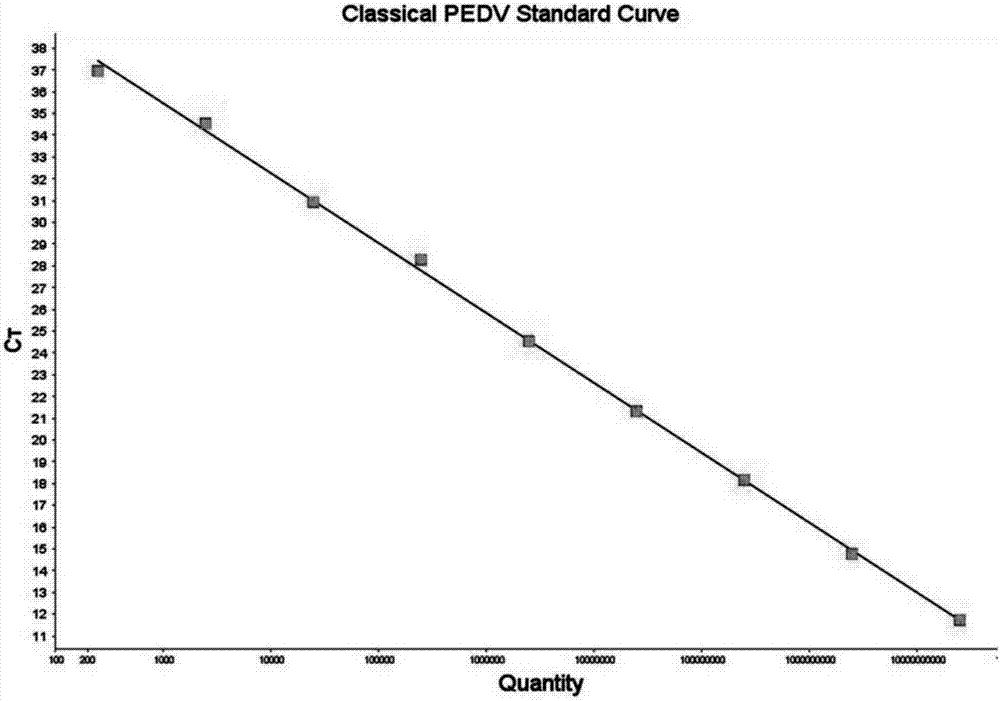
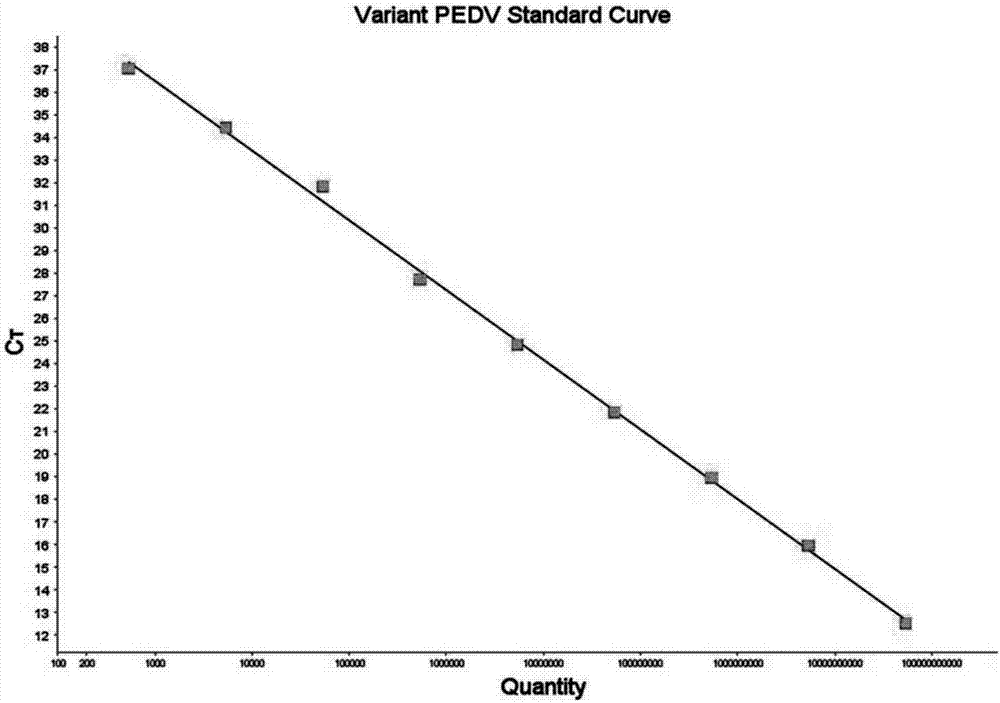
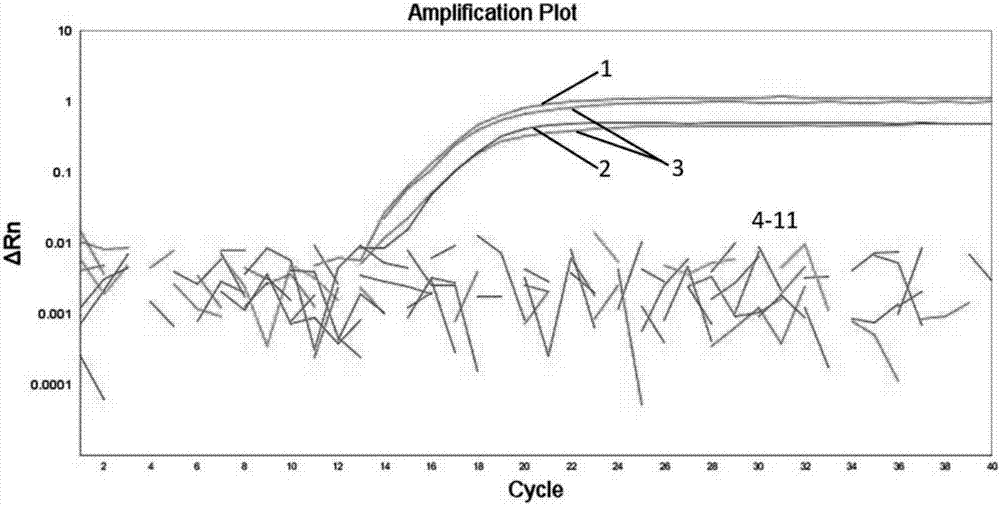
Porcine epidemic diarrhea virus (PEDV) fluorescent quantitative PCR primer and probe
A porcine epidemic diarrhea and fluorescence quantitative technology, applied in the field of molecular biology, can solve the problem of undetected mutant strains, and achieve the effects of fast detection speed, simple operation and high accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Example 1: Design and synthesis of probes and primers
[0047] The primers and probes are the classic strains of porcine epidemic diarrhea virus published in GenBank in recent years by the DNAMAN8 software (the accession numbers of the representative strains of the classic strains are: AF353511.1, JN547228.1, KT323979.1, EF185992. 1, JN601050.1, JX560761.1) and mutant strains (the accession numbers of the mutant strains are: KC210145.1, JX112709.1, KU133250.1, KJ646584.1, KP698764.2, KT428879.1, KX073609.1, KU133236.1, KU133259.1, KU237226.1, KU710232.1, KR233257.1, KJ646580.1, KU237230.1, KU237229.1, KU977512.1, KU710235.16U71.2K2, KU133 1, KU710234.1, KU710229.1, KU710242.1, KR296683.1, KT989356.1, KU237217.1) S1 gene sequence alignment, design two specific probes in the sequence difference region, and at the probe position Design common primers for the periphery, and then use MFEprimer-2.0 to evaluate the feasibility of probes and primers. All primers and probes we...
Embodiment 2
[0048] Example 2: RNA extraction and cDNA synthesis
[0049] Take 300 μl of the cell culture medium infected with the classic strain of porcine epidemic diarrhea virus CV777 (this strain has been identified as a classic strain of porcine epidemic diarrhea virus, and other classic strains can also be used instead), 300 μl of the cell culture medium infected with the variant virus of porcine epidemic diarrhea virus Strain CH / HNLY (this strain has been identified as a porcine epidemic diarrhea virus variant strain, and other variant strains can also be used instead) clinical sample processing supernatant (take the infected porcine epidemic diarrhea virus variant strain CH / HNLY Cut pig intestinal tissue into pieces with scissors, add appropriate amount of normal saline, freeze-thaw three times, centrifuge at 3000rpm / min for 5min to get the supernatant), add 800μl Trizol respectively, mix well, and let stand at room temperature for 10min; then add 200μl chloroform, fully Mix well, ...
Embodiment 3
[0051] Embodiment 3: gene cloning
[0052] Using the cDNA obtained by reverse transcription as a template, use the upstream and downstream primers (SEQ ID NO.1-SEQ ID NO.2) of the present invention to amplify the target fragment respectively, wherein the PCR amplification reaction system is: template 1 μl, 2× Ex Taq 10μl, upstream and downstream primers (10pmol) each 1μl, supplemented with deionized water to 20μl; PCR reaction program: 95°C pre-denaturation for 5min, and then the following 30 cycles: 95°C denaturation for 30s, 55°C for 30s, 72°C Extend for 30s, and then extend for 5min at 72°C after the cycle ends. The PCR amplified product was identified by 1% agarose electrophoresis, and the target fragment was recovered with a gel recovery kit (TaKaRa), and the target fragment was cloned into the Peasy-Blunt vector and transformed into DH5α competent cells. The positive clones were picked by blue-white screening and sequenced. Positive recombinant plasmids were identified....
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com