Kit for rapidly detecting nucleic acid of hepatitis C virus and detection method of kit
A hepatitis C virus and kit technology, applied in the field of biomedical detection, can solve the problems of low detection throughput, difficulty in rapid amplification, etc., and achieve the effects of high sensitivity, easy melting and denaturation, and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Embodiment 1, the preparation of kit
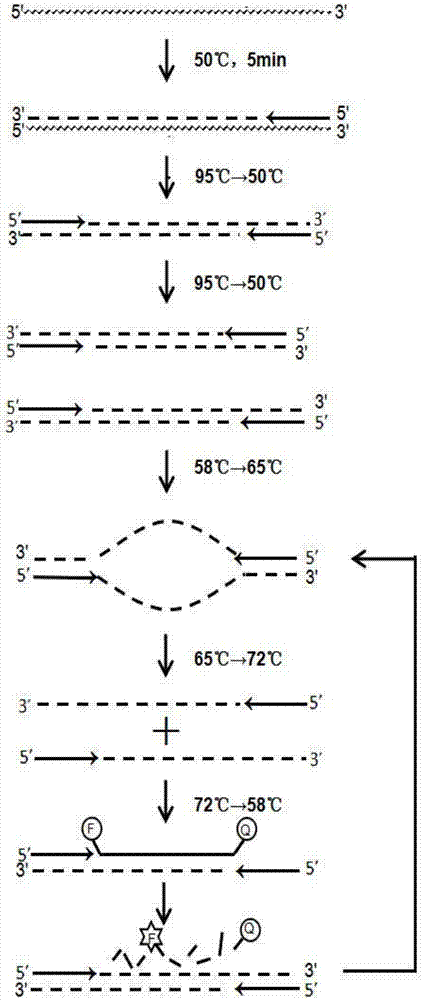
[0047] The kit for rapid detection of hepatitis C virus nucleic acid using one-step fluorescent quantitative RT-qPCR technology of the present invention includes nucleic acid extraction liquid, RT-qPCR reaction liquid, enzyme mixed liquid, negative control, positive control, and positive reference product.
[0048] 1. Based on the sequence information of the hepatitis C virus genome database of different genotypes, use Clustal Omega software (http: / / www.ebi.ac.uk / Tools / msa / clustalo / ) to do multiple sequence comparison analysis to find different genes The conserved region of type, design the primer sequence and probe sequence that are used for the specific detection of hepatitis C virus nucleic acid, and do BLST comparison, finally artificial synthesis above-mentioned designed primer and probe, its nucleotide sequence is respectively:
[0049] Upstream primer: 5'-ACTGCTAGCCGAGTAGTGTTG-3' (SEQ ID NO.1);
[0050] Downstream primer: 5...
Embodiment 2
[0076] Embodiment 2, the use of kit
[0077] 1. Sample processing: centrifuge the blood to be tested at 1500g for 20 minutes, discard the lower blood cells, transfer the upper yellow liquid to a new storage tube, and store it at -20°C for later use.
[0078] 2. HCV RNA extraction
[0079] 1) Take 200 μl of the above samples, positive control, negative control and positive reference respectively into 1.5ml centrifuge tubes, and mark them well.
[0080] 2) Add 50 μl proteinase K and 20 μl magnetic microspheres to the centrifuge tube.
[0081] 3) Add 400 μl of RNA extraction solution I to each centrifuge tube, and incubate with shaking at room temperature for 5 minutes.
[0082] 4) Place the centrifuge tube on the magnetic stand and let it stand for 2 minutes. When the magnetic beads are completely adsorbed, carefully remove the liquid.
[0083] 5) Remove the centrifuge tube from the magnetic stand, add 600 μl RNA extraction solution II, shake and mix for 30 seconds.
[0084]...
Embodiment 3
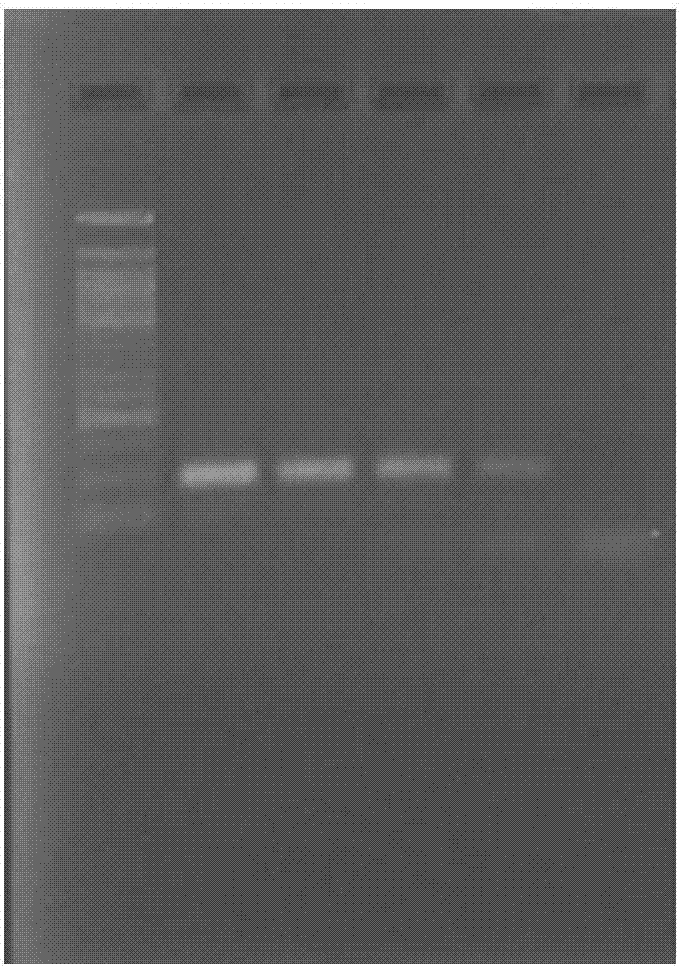
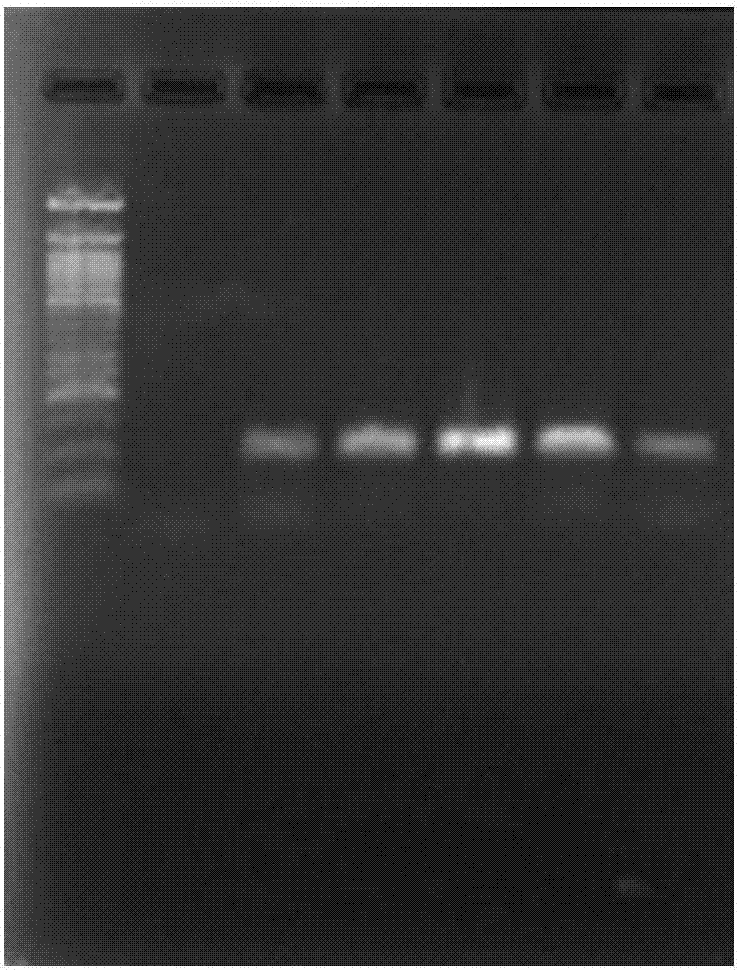
[0096] Embodiment 3, under different temperatures, the impact of denaturant A (5%) on the amplification efficiency of hepatitis C virus nucleic acid
[0097] According to the method of using the above kit, use the hepatitis C virus nucleic acid positive sample (1000IU / ml) as a template to extract nucleic acid RNA, RT-PCR amplification, and the fluorescent quantitative PCR reaction program is only set in the third part (fast part) ) change the denaturation temperature, respectively: 95°C (swimming lane 2), 90°C (swimming lane 3), 85°C (swimming lane 4), 80°C (swimming lane 5) and 75°C (swimming lane 6). Agarose gel electrophoresis separation, staining (EB), the results are as follows figure 2 shown. It can be clearly seen from the figure that the addition of denaturing agent A (5%) can reduce the denaturation temperature, wherein, in the range of 85-95°C, the amplification can be well performed without significant difference, while at 80°C, although the denaturation temperatu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com